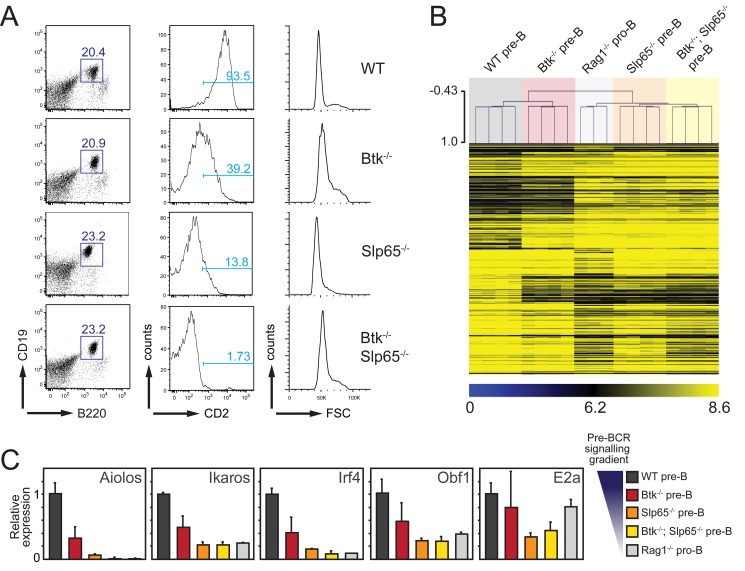
Figure 1. Gene expression profiling strategy for the identification of genes regulated by Btk/Slp65-mediated pre-BCR signaling.
(A) FACS sorting strategy for purification of pre-B cell fractions from the indicated mice on a VH81x transgenic Rag1 −/− background. Lymphocytes were gated on the basis of forward/side scatter and B220+CD19+ pre-B cell fractions were sorted. Virtually all B220+CD19+ cells were cytoplasmic μ heavy chain positive [34], but showed genotype-dependent levels of expression of the CD2 differentiation marker, in agreement with previous findings [34]. (B) DNA microarray analysis of total mRNA from FACS-purified B220+CD19+ pre-B/pro-B cell fractions from the indicated mice. One-way ANOVA analysis (p = 0.01) identified 266 significantly different genes. MeV hierarchical clustering of gene expression differences are represented in the heatmap. (C) Validation of the expression of TFs implicated in Igκ gene rearrangement. Total mRNA isolated from FACS-sorted B220+CD19+ pre-B/pro-B cell fractions from the indicated mice was analyzed by quantitative RT-PCR for expression of TFs. Expression levels were normalized to those of Gapdh, whereby the values in WT pre-B cells were set to one. Bars represent mean values and error bars denote standard deviations for four independent mice per group.