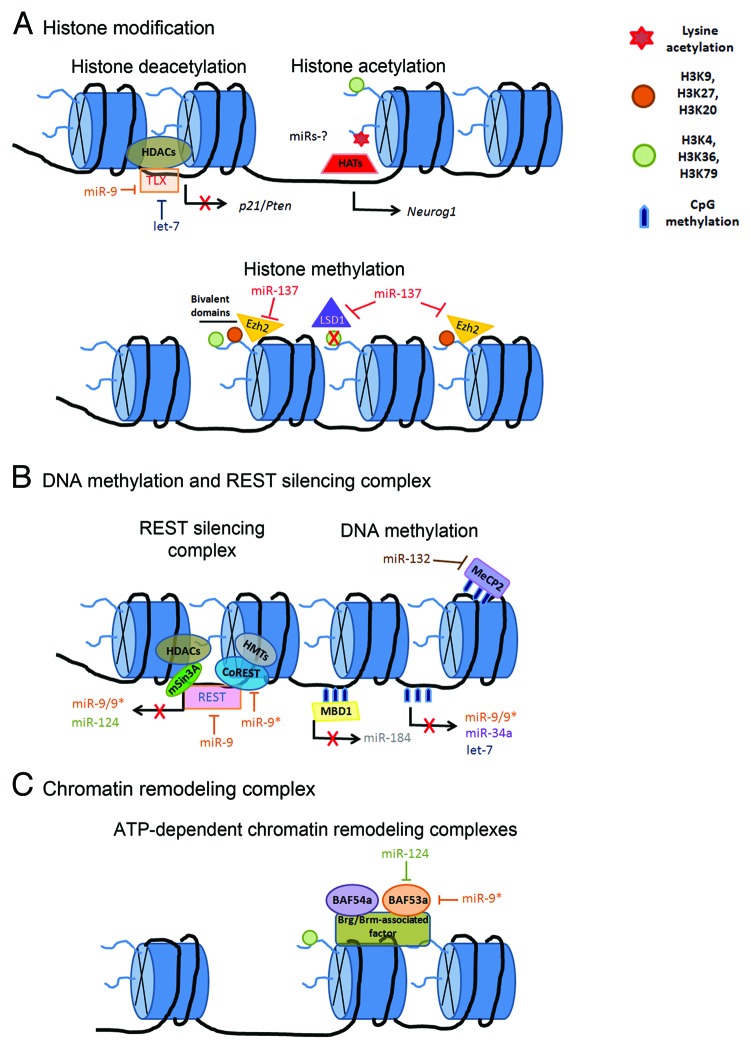
Figure 2. Molecular mechanisms of the epigenetic-miRNAs regulatory network associated with chromatin remodeling during neurogenesis. (A) Histone modifications are mediated by acetylation, deacetylation and methylation. HATs relax chromatin structure and facilitate transcription, while HDACs increase chromatin condensation and repress transcription. miR-9 and let-7 regulate the expression of the transcription factor TLX that directly recruits HDACs in order to mediate p21 and Pten gene repression in NSCs. miR-137 has emerged as an important regulator of histone methylation regulatory pathway by targeting the demethylase, LSD1, and the H3K27 methyltransferase, Ezh2. miR-137-Ezh2 network might be associated in balancing gene-specific bivalent domains in NSCs.2 (B) DNA methylation is associated with transcriptional repression and negative regulation of several brain-enriched miRNAs. The transcriptional repressor MBD protein, MeCP2, binds to methylated genes in order to silence genes. MeCP2 has been identified as a target of miR-132 in neurons. The transcription factor REST regulates the expression of brain-enriched miR-9/9* and miR-124. miR-9/9* act as a negative feedback regulatory loop on REST silencing complex. (C) ATP-dependent chromatin remodeling complex alter the chromatin structure in order to regulate NSC fate determination. BAF53a expression levels are regulated by miR-124 and miR-9* during the transition between NSCs proliferation to differentiated post-mitotic neuron.122

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.