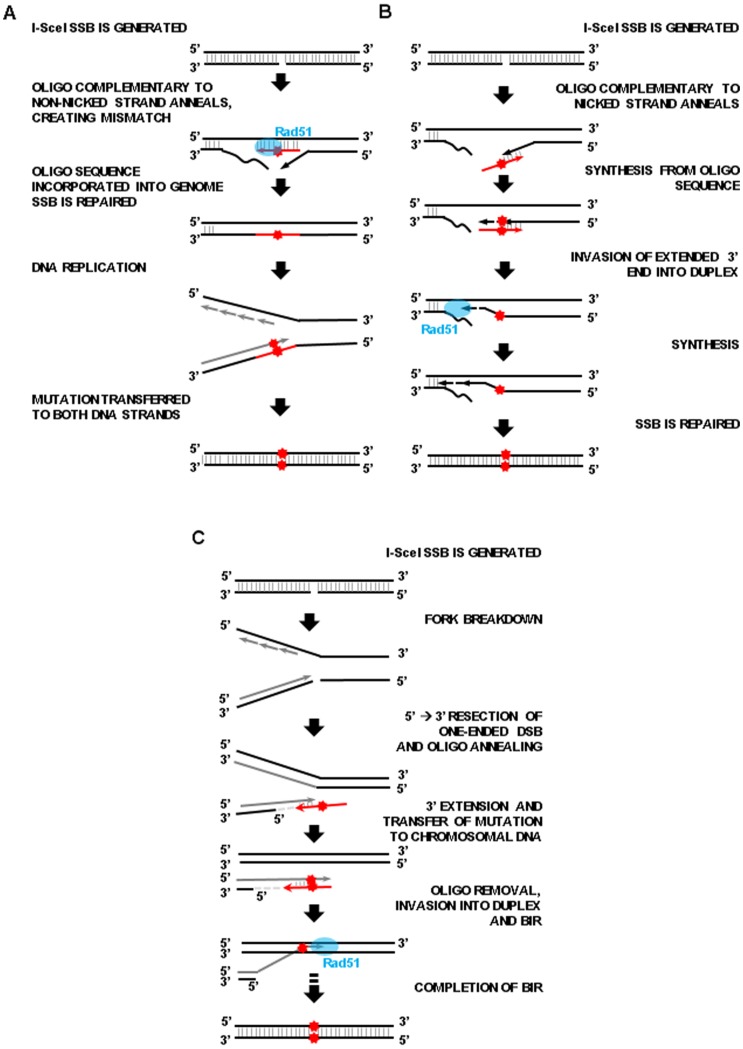
Figure 8. Models for I-SceI K223I SSB-driven HR using single-stranded oligonucleotides.
(A) Gene correction using an oligonucleotide complementary to the intact strand. After the SSB is generated, an oligonucleotide (red arrow) carrying the desired nucleotide change (red star) anneals to the complementary strand by invading the nicked duplex with the help of Rad51. The oligonucleotide is incorporated in the duplex and its genetic modification is transferred to the other strand in the subsequent round of DNA replication. (B) Gene correction using an oligonucleotide complementary to the nicked strand. After the SSB is generated, an oligonucleotide sequence (red arrow) with the desired mutation (red star) can serve as template to extend the unwound 3′ broken end. This extended 3′ end may invade the duplex via Rad51 function. The 3′ end is then extended further. The nick is repaired and the mutation fixed by mismatch repair or in the next round of replication. (C) Gene correction following collapse of a replication fork at the nick. After the SSB is generated, it persists until encountered by the replication fork. Following fork breakdown, a one-ended DSB forms. Resection of the 5′ end (indicated by a light gray dashed line) produces a single-strand 3′ end that anneals with the complementary sequence of an oligonucleotide (red arrow) containing a desired mutation (red star) and uses the oligonucleotide as a template for extension. After removal of the oligonucleotide by unwinding or resection, the 3′ end (dark gray) invades the intact duplex via Rad51 and follows the steps (dotted black arrow) of BIR to complete repair. The mutation carried on the 3′end is passed to the other strand of the chromosome by mismatch repair or in the next round of replication.