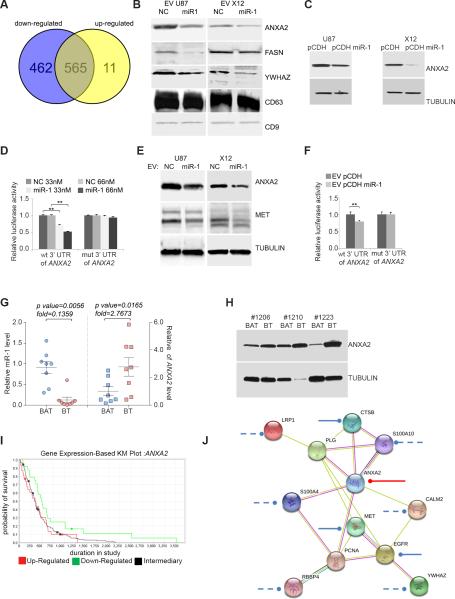
FIGURE 6. MiR-1 directly targets EV ANXA2 overexpressed in human GBM.
A. Venn diagram depicting differential protein composition of EV's derived from U87 cells stably infected with pCDH-GFP miR-1 expressing vector (downregulated: blue, upregulated: yellow) compared to EVs derived from U87 cells stably infected with pCDH-GFP control vector.
B. EV proteins were validated by Western blotting analysis. EVs were derived from U87 and X12 cells lines stably infected with either pCDH-GFP control vector (pCDH) or pCDH-GFP miR-1 expressing vector (pCDH miR-1). EV lysates were blotted with specific antibodies against indicated proteins. CD9 and CD63 were used as a loading control.
C. Effects of miR-1 on the expression of ANXA2 were validated by Western blotting analysis. U87 and X12 GBM cell lines were stably expressing pCDH-GFP control vector (pCDH) or pCDH-GFP miR-1 expressing vector (pCDH miR-1). Cell lysates were blotted with antibodies against ANXA2. Tubulin was used as a loading control.
D. Direct targeting of ANXA2 3′UTR by miR-1 was validated using a luciferase/3’UTR reporter assay. COS7 cells were co-transfected with luciferase/ANXA2 wild type 3′UTR reporter vector (wt) and33 nM or 66 nM negative control miR (NC) or miR-1. A reporter vector with a mutated miR-1 binding site in the ANXA2 3′UTR (mut) was used as a control. Luciferase levels are expressed as mean relative to controls ± SD; **P < 0.01.
E. Effects of EV-carried miR-1 on the expression of ANXA2 and MET were validated by Western blotting analysis. U87 and X12 cells were exposed to the presence of EVs derived from corresponding cells transiently transfected with either negative control miR (NC) or miR-1 oligonucleotides. Cell lysates were blotted with antibodies against ANXA2, MET and tubulin as a loading control.
F. Direct targeting of ANXA2 3′UTR by EV-carried miR-1 was validated using luciferase/3’UTR reporter assay. U87 and X12 cells were exposed to the presence of EVs derived from corresponding cells transiently transfected with either negative control miR (NC) or miR-1 and after 24h transfected with luciferase/ANXA2 3′UTR wild type (wt) and mutant (mut) reporter vector. Luciferase levels are expressed as mean relative to controls ± SD; **P < 0.01.
G. Relative expression of miR-1 (left) and ANXA2 mRNA (right) levels were validated by qPCR in GBM brain tumor (BT) specimens vs. matching brain adjacent to tumor (BAT) (n = 8). Values are expressed as mean relative miR-1 expression level ± SD.
H. ANXA2 protein level was validated in GBM by Western blotting analysis. Cell lysates from GBM brain tumor (BT) specimens vs. matching brain adjacent to tumor (BAT) (n = 3) were blotted with antibodies against ANXA2. Tubulin was used as a loading control.
I. Association of ANXA2 expression with patient survival (Kaplan–Meier [KM] plot) in GBM. Data was obtained from The Cancer Genome Atlas. Upregulated (n = 31), downregulated (n = 24), and intermediary samples (n = 156) were analyzed. Up- vs. downregulated P = 0.05; up-regulated vs. intermediary P = 0.7134; down-regulated vs. intermediary P = 0.0147.
J. MiR-1 dependent targeting of EV ANXA2 network. ANXA2 partners were selected from EV proteins carried differentially in miR-1-dependent manner. Experimentally validated miR-1 targeting of ANXA2 is shown as a red line; targeting based on published data are shown as a solid blue lines, putative targeting based on target prediction software only are shown as a dashed lines. Networking was analyzed with STRING software.