Figure 1.
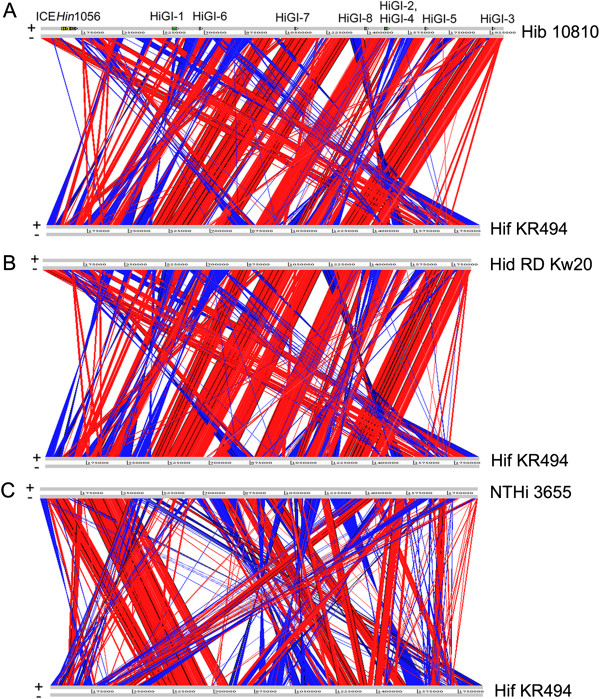
Genome comparison of H. influenzae type f KR494 and reference strains in ACT view. Genome alignment of Hif KR494 and (A) H. influenzae type b (Hib) 10810, (B) type d (Hid) Rd Kw20, and (C) nontypeable H. influenzae (NTHi) 3655. Respective genome designations are indicated on the right hand side of each genome line. Forward (+) and complement (−) strands of each genome are indicated in gray genome lines. Genomes are shown in full length and drawn to scale. Direct and inverted synteny between individual ORF (not indicated here) of the comparing genomes are shown in red and blue, respectively. The level of amino acid similarity is represented by colour shading with ascending saturation and indicates higher similarity. Genetic islands (HiGi) and the ICE element identified from Hib strain Eagan and strain 1056, respectively, are indicated for Hib 10810 in the upper genome line in panel (A).
