Figure 3.
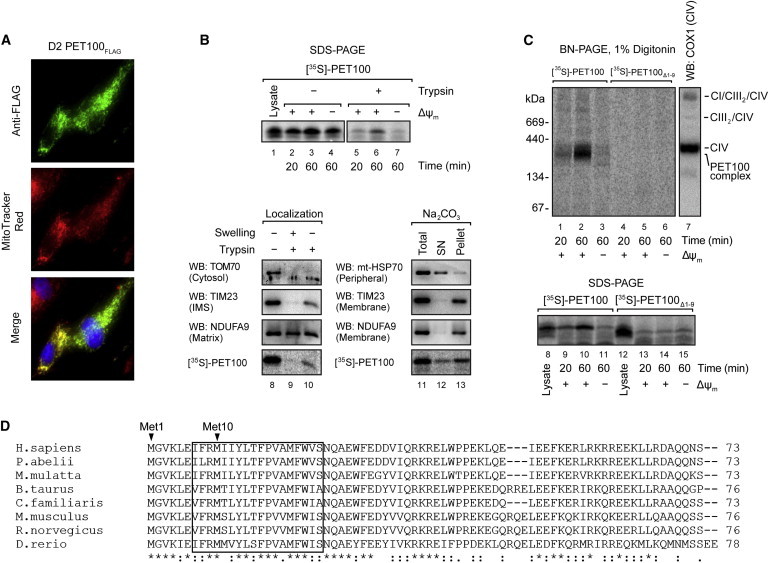
Mitochondrial Localization and Assembly of PET100
(A) D2 fibroblasts expressing PET100FLAG were stained with MitoTracker Red (red), fixed, and immunostained for the FLAG epitope (green). Nuclei were labeled with Hoechst (blue). Colocalization of the expressed PET100FLAG in the mitochondria (yellow) is shown in the merged image.
(B) Top: The presence of membrane potential (ΔΨm) was required for the import of 35S-labeled PET100 into isolated HEK293T mitochondria, protecting it from digestion by externally added trypsin. Untreated input lysate is shown for comparison. 10% of input lysate used in the import was loaded in lane 1. Bottom: [35S]PET100 imported into HEK293T mitochondria was digested by trypsin only after hypo-osmotic mitochondrial swelling (left) and remained in the membrane (Pellet) fraction after alkaline extraction (Na2CO3) (right). Abbreviations are as follows: SN, supernatant; WB, immunoblotting.
(C) BN-PAGE (top) or SDS-PAGE (bottom) followed by phosphorimaging showed that [35S]PET100Δ1-9 was incapable of assembly into the ∼300 kDa complex (lanes 4–6) and was not imported into mitochondria (lanes 13–15). Abbreviations are as follows: CI, complex I; CIII, complex III; CIV, complex IV.
(D) Sequence alignment of human PET100 with its homologs in seven additional vertebrate species. The PET100 in the Lebanese LS individuals (if present) was predicted to lack the first nine amino acid residues, which are highly conserved in vertebrate species. Asterisk (∗), colon (:), and period (.) indicate that the amino acids are identical, strongly similar, and weakly similar, respectively, across the aligned species. The transmembrane domain predicted from the human protein67 is boxed.
