Figure 1. Library design using the Oligopaints strategy.
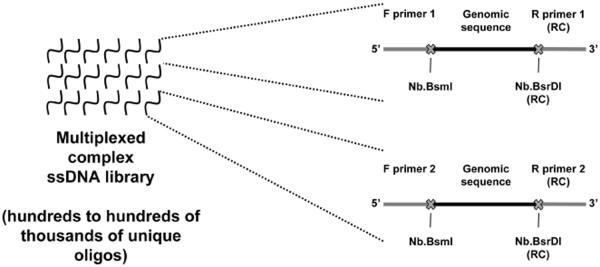
Each synthetic ssDNA library may contain hundreds to hundreds of thousands or more unique oligo species and can contain multiple distinct Oligopaint probes through the use of multiple primer pairs. This figure diagrams the structure of two oligos from different probe sets in a hypothetical library: an oligo belonging to a probe that can be amplified using primer pair 1 (top left) and an oligo belonging to a probe that can be amplified using primer pair 2 (bottom left). Both oligos contain, in 5' to 3' order, the sequence of the forward primer, genomic sequence, and then the reverse complement (RC) of the reverse primer. The sites for two nicking endonucleases, Nb.BsmI and Nb.BsrDI, are placed between the primer sequences and the genomic sequences. The use of two nicking endonuclease sites in the library molecules allows for the production of strand specific probes: amplification with a labeled F primer and digestion with Nb.BsrDI will yield probe targeting the reverse complement of the genomic sequence encoded in the ssDNA molecules of the library, whereas amplification with a labeled R primer and digestion with Nb.BsmI will yield probe targeting the genomic sequence encoded in the ssDNA molecules of the library. The use of two nicking endonuclease sites, instead of one, is an update of the strategy presented in Figure 1 of Beliveau et al. (2012).
