Figure 2. examples of FISH with Oligopaint probes.
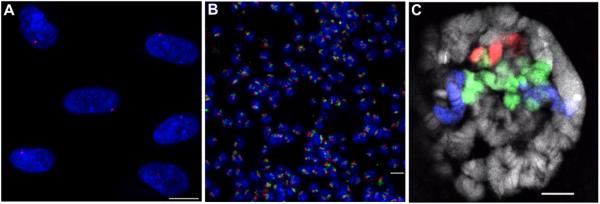
(A) FISH using an Oligopaint probe composed of 20,020 oligos targeting a 2.5 Mb region on human chromosome X (Xq13.1) in diploid (XY) MRC-5 cells. Hybridization was carried out according to Basic Protocol 2 in a 25 μl volume using 35 pmol of probe (Cy3, red) produced using Basic Protocol 1. DNA is stained with DAPI (blue). (B) Two-color FISH on a field of tetraploid Drosophila Kc167 cells using two Oligopaint probes targeting the right arm of chromosome 2: one composed of 25,000 oligos targeting a 2.7 Mb region (50D1-53D7), and another composed of 50,000 oligos - half targeting a 3 Mb region (41E3-44C4) and the other half targeting a 2.6 Mb region (58D2-60D14). The hybridization was carried out according to Basic Protocol 2 in a 25 μl volume using 5 pmol of the first probe (Cy3, red) and 2.5 pmol of the second probe (Cy5, green), both produced using Basic Protocol 1. (C) Three-color FISH performed on Drosophila salivary polytene chromosomes with the two Oligopaint probes used in (B) plus an additional Oligopaint probe that also targets the right arm of chromosome 2 and is composed of 105,000 oligos - half targeting a 5.6 Mb region (44C4-50C9) and the other half targeting a 5.5 Mb region (53C9-58B6). The hybridization was carried out according to Beliveau et al. (2012) in a 25 μl volume using 20 pmol of each of the three probes labeled with TYE563 (red), TYE665 (blue), and 6-FAM (green), respectively, all produced using Basic Protocol 1. DNA is stained with DAPI (gray). Images represent confocal maximum Z projections (A, B) or a single confocal XY plane (C). Scale bars: 10 μM.
