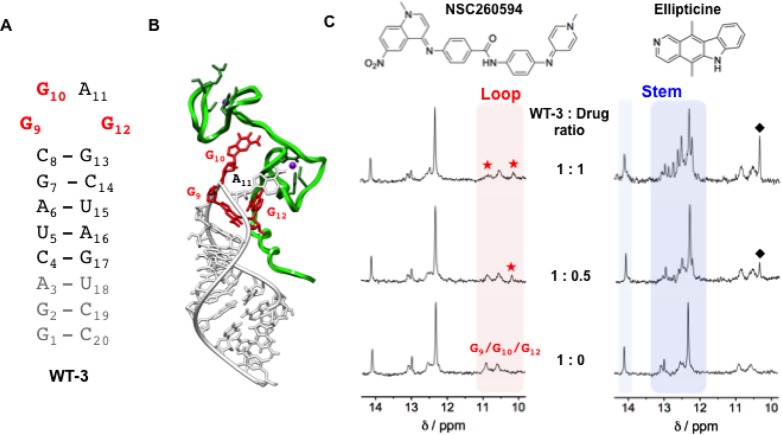
Figure 3.
Interaction between the structural analogue of SL3 (WT-3) and the two small molecules, NSC260594 and ellipticine, as monitored by 1H NMR. (A) Secondary structures of the WT-3 hairpin. In gray are reported the nucleotides added to SL3 in order to have a well-defined secondary structure suitable for NMR spectroscopy and in red the G9, G10, and G12 guanines constituting the tetraloop of SL3. (B) 3-D model of the HIV-1 nucleocapsid–WT-3 complex (PDB 1A1T) depicting the protein and the RNA hairpin with a green and white ribbon, respectively. The G9, G10, and G12 guanines essential for binding to the HIV-1 nucleocapsid are represented in red. (C) WT-3 1H NMR titration experiments with an increasing amount of the compounds NSC260594 and ellipticine. The imino protons between 12 and 14.5 ppm (blue region) are attributed to the Watson–Crick H-bonded base pairs of the stem of the hairpin structure and imino protons between 10 and 11 ppm (red region) are attributed to the WT-3 loop G bases. Red stars (★) highlight the alteration of the G9, G10, and G12 imino signals, and black diamonds (⧫) highlight peaks attributed to ellipticine.