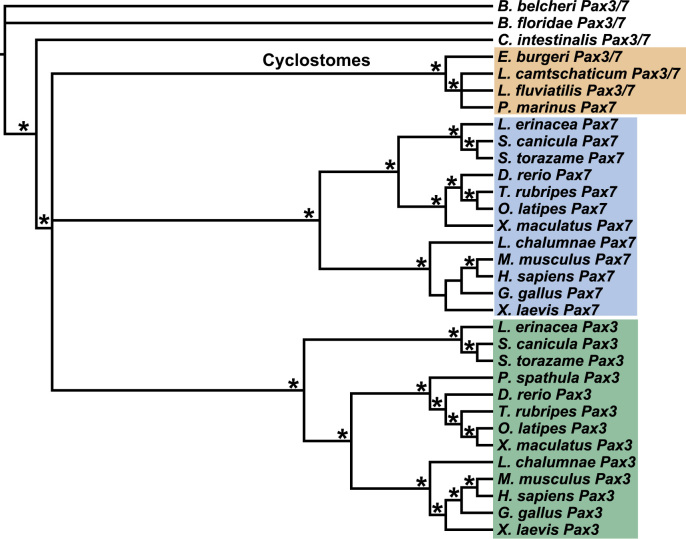
Fig. 2.
Cyclostome Pax3/7 subfamily proteins form a well-supported clade, separate from gnathostome Pax3 and Pax7 clades. Maximum a posteriori (MAP) topology, obtained with Bali-Phy software, summarizing all of the phylogenetic analyses performed. Nodes consistently recovered with high support in all Bayesian and coalescence-based analyses are indicated by an asterisk (where the posterior probability is >90). These analyses consistently recovered three main clades of orthologous sequences: (i) a cyclostome clade containing all published hagfish and lamprey Pax3/7 proteins (orange box), (ii) a gnathostome Pax7 clade (blue box) and (iii) a gnathostome Pax3 clade (green box). Relationships between these three clades, however, remain unresolved: the gnathostome Pax3 and Pax7 clades formed a sister group in most analyses but with low support. Within the gnathostome clades, relationships are well resolved for the Pax3 but not the Pax7 subfamily: in some cases, short sequence lengths for Pax7 likely account for a topology that is incongruent with the currently proposed relationships among vertebrates. Generally, however, phylogenetic relationships recovered here are consistent with current understanding of the chordate phylogeny.