Figure 3. N-Myc represses RD3 gene transcription by direct binding to the RD3 gene promoter.
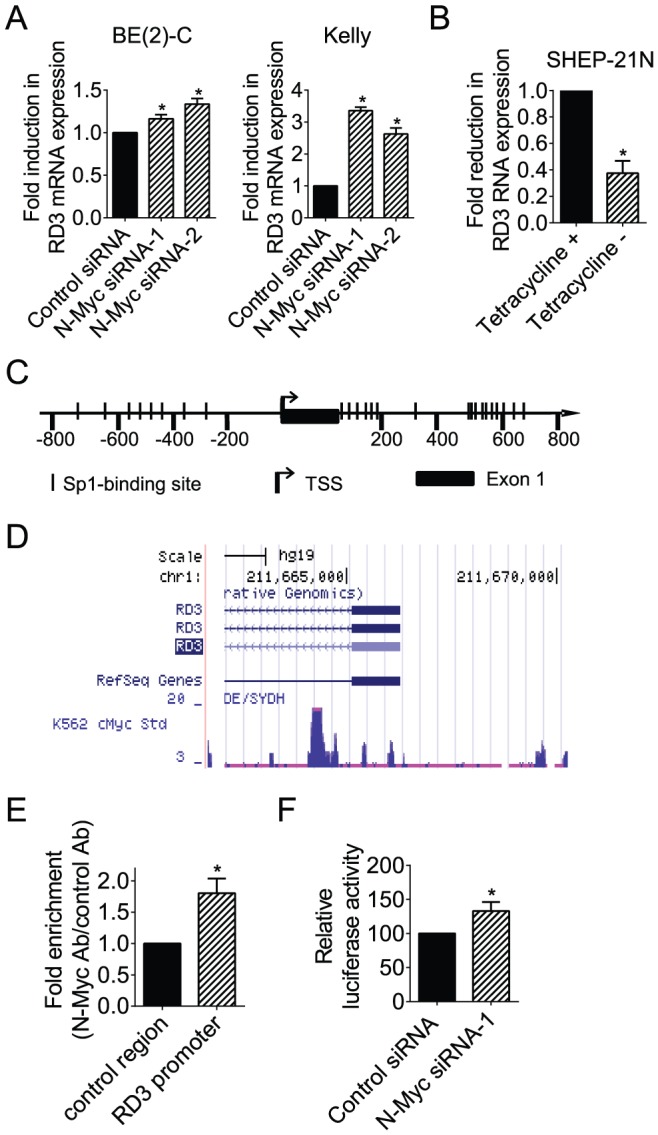
(A) BE(2)-C and Kelly cells were transfected with scrambled control (Cont) siRNA, N-Myc siRNA-1 or N-Myc siRNA-2 for 48 hours, followed by RNA extraction and real-time RT-PCR analyses of RD3 mRNA expression. (B) SHEP-21N cells were incubated with or without tetracycline for 48 hours, followed by RNA extraction and RT-PCR analysis of RD3 RNA expression. (C) Schematic representation of the RD3 gene promoter. TSS represented transcription start site, and | represented Sp1-binding sites. (D) ChIP-Seq data from Dr. Michael Snyder's group at Yale University for the ENCODE/SYDH project generated from K562 cells. (E) ChIP assays were performed with a control or anti-N-Myc antibody (Ab) and primers targeting a negative control region or the RD3 gene core promoter region enriched in Sp1-binding sites in BE(2)-C cells. Fold enrichment was calculated by dividing PCR products from DNA samples immunoprecipitated with the anti-N-Myc Ab by PCR products from DNA samples immunoprecipitated with the control Ab, relative to input. Fold enrichment at the negative control region was artificially set as 1.0. (F) BE(2)-C cells were transfected with control siRNA or N-Myc siRNA-1, followed by co-transfection with Cypridina TK control construct plus empty vector or RD3 gene promoter pLightSwitch_Prom construct. Luciferase activities were measured with a LightSwitch Dual Assay System kit, and expressed as a percentage change relative to control siRNA transfected samples. Error bars represented standard error. * indicated P<0.05.
