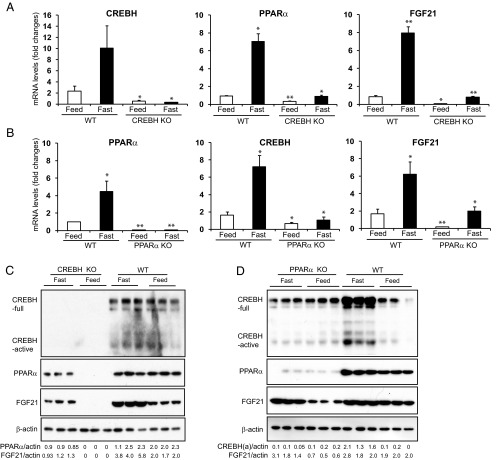
Figure 2.
Fasting stress induces an interdependent regulatory axis of CREBH-PPARα-FGF21 in the liver. A and B, Expression levels of CREBH, PPARα, and FGF21 mRNAs in the livers of wild-type (WT) control, CREBH-null (A), and PPARα-null (B) mice under feeding or fasting condition. Total RNAs were isolated from liver tissues of WT, CREBH-null, and PPARα-null mice under the normal chow diet (feeding) or after 14 hours of fasting. These RNAs were subjected to quantitative real-time RT-PCR analysis. Expression values were normalized to β-actin mRNA levels. Fold changes of mRNA levels are shown by comparison to that in one of the WT control mice under the feeding condition. Each bar denotes the mean ± SEM (n = 3). *, P < .05; **, P < .01.WT, wild-type; KO, knockout. C and D, Levels of the CREBH, PPARα, and FGF21 proteins in the livers of WT control, CREBH-null (C), and PPARα-null (D) mice under feeding or fasting condition. Liver whole lysates were prepared from the WT, CREBH-null, or PPARα-null mice under the normal chow (feeding) or after 14 hours of fasting. Equal amounts of the proteins were separated on SDS-PAGE gels and immunoblotted using the antibody against CREBH, PPARα, FGF21, or β-actin. The values below the gels represent the PPARα, FGF21, or activated form of CREBH protein signal intensities that were quantified using NIH ImageJ software and normalized to β-actin signal intensities.