Abstract
Background:
Jumonji domain-containing protein 2B (JMJD2B), directly targeted by hypoxia-inducible factor 1α, maintains the histone methylation balance important for the transcriptional activation of many oncogenes. Jumonji domain-containing protein 2B has been implicated in colorectal cancer (CRC) progression; however, the mechanism remains unclear.
Methods:
Immunofluorescence and western blotting detected phosphorylated histone H2AX, characteristic of double-strand breaks, and comet assay was used to investigate DNA damage, in CRC cells after JMJD2B small interfering RNA (siRNA) transfection. We assessed the resulting in vitro responses, that is, cell cycle progression, apoptosis, and senescence coupled with JMJD2B silencing-induced DNA damage, studying the regulatory role of signal transducers and activators of transcription 3 (STAT3). The JMJD2B silencing anti-cancer effect was determined using an in vivo CRC xenograft model.
Results:
Jumonji domain-containing protein 2B knockdown induced DNA damage via ataxia telangiectasia-mutated (ATM) and ATM and Rad3-related pathway activation, resulting in cell cycle arrest, apoptosis, and senescence in both normoxia and hypoxia. Signal transducers and activators of transcription 3 suppression by JMJD2B silencing enhanced DNA damage. Intratumoural injection of JMJD2B siRNA suppressed tumour growth in vivo and activated the DNA damage response (DDR).
Conclusions:
Jumonji domain-containing protein 2B has an essential role in cancer cell survival and tumour growth via DDR mediation, which STAT3 partially regulates, suggesting that JMJD2B is a potential anti-cancer target.
Keywords: JMJD2B, DNA damage response, hypoxia, STAT3, colorectal cancer
Emerging evidence indicates that hypoxia is a major component of human cancer progression and induces a range of cellular responses, especially epigenetic responses (Watson et al, 2010), of which histone methylation status imbalances because of dysregulated histone methyltransferases and demethylases have been linked to cancer (Shi, 2007; Zeng et al, 2010). Jumonji domain-containing protein 2B (JMJD2B) is a newly identified member of the histone demethylase JMJD2 family that is characterised by the catalytic Jumonji C domain. Jumonji domain-containing protein 2B specifically targets the trimethylated lysine 9 of histone H3 (H3K9me3) for demethylation at the pericentric heterochromatin and euchromatin (Fodor et al, 2006; Shi et al, 2011). Expression levels of JMJD2B are notably upregulated in numerous cancers, including breast cancer (Yang et al, 2010; Kawazu et al, 2011; Shi et al, 2011), gastric cancer (Li et al, 2011; Kim et al, 2012), and colorectal cancer (CRC; Fu et al, 2012). Jumonji domain-containing protein 2B also has a crucial role in human carcinogenesis by regulating cell cycle progression and apoptosis (Toyokawa et al, 2011; Zhu et al, 2012; Wen et al, 2012). Recently, we reported that JMJD2B upregulates the hypoxia-inducible genes involved in cancer cell proliferation and invasion by specifically demethylating the promoter-repressive H3K9me3 (Fu et al, 2012). However, little is currently known about the other cellular mechanisms of action of JMJD2B.
Recent work has revealed the intricate alterations in histones, accompanied by the DNA damage response (DDR), which is necessary for facilitating signalling, checkpoints, and DNA repair. Many tumour cells exhibit increased chromosome instability, which has been proven to be linked with the loss of H3K9me3 because of JMJD2B overexpression in breast cancer (Slee et al, 2012). Hypoxia can induce genetic instability, which is associated with the dysregulation of many DNA repair pathways, such as DNA mismatch repair (Bindra and Glazer, 2007a; Rodríguez-Jiménez et al, 2008), nucleotide excision repair (Yuan et al, 2000), and double-stranded break (DSB) repair (Bindra et al, 2005; Chan et al, 2008). As JMJD2B is regulated by hypoxia-inducible factor 1 α (HIF-1α), it may also be involved in the DDR leading to genetic instability in cancer. Furthermore, JMJD2A inhibition combined with JMJD2B can facilitate the recruitment of p53-binding protein 1 to DNA damage sites (Mallette et al, 2012); Drosophila cells expressing JMJD2B mutants are more sensitive to ultraviolet-induced DNA damage (Palomera-Sanchez et al, 2010).
In light of these studies indicating the importance of JMJD2B to cancer cell growth and genetic integrity, we investigated whether CRC cell proliferation and survival are associated with the DDR upon JMJD2B silencing by small interfering RNA (siRNA). We found that JMJD2B suppression remarkably stimulated the DDR, and consequently induced tumour cell cycle arrest, apoptosis, and senescence, which is partially regulated by signal transducers and activators of transcription 3 (STAT3). This suggests that JMJD2B possesses multiple functions, apart from its traditional role as a histone demethylase, and is a potential anti-cancer target.
Materials and Methods
Cell culture
The human CRC cell lines HCT116 and SW480 were cultured at 37 °C in 95% air, 5% CO2 in McCoy's 5A, and RPMI-1640 media (Gibco BRL, Gaithersburg, MD, USA) supplemented with antibiotics and 10% fetal bovine serum. For hypoxia treatment, cells were incubated in 1% O2, 5% CO2, and 94% N2 in a hypoxia chamber (YCP-50S; Huaxi Electronic Technologies, Changsha, China).
SiRNA transfection
SiRNAs were transfected into subconfluent cells using Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The siRNAs specifically targeting HIF-1α, JMJD2B, and STAT3 have been described previously (Du et al, 2012; Fu et al, 2012). The specificity of JMJD2B siRNA has been examined (Supplementary Figure 1). Non-targeting control siRNA was purchased from GenePharma (Shanghai, China). Hypoxia-inducible factor 1 α and STAT3 siRNA transfections were carried out in 20% confluent cells for 48 and 24 h, respectively, before 24 h hypoxia treatment. For JMJD2B silencing, 20% confluent CRC cells were transfected with JMJD2B siRNA for 24 h and then underwent 0, 6, 12, or 24 h hypoxia treatment.
Plasmids transfection
Full-length STAT3 and JMJD2B cDNA were obtained by PCR from a human cDNA library. To construct the eukaryotic expression vectors, the STAT3 and JMJD2B cDNA were cloned into a pCDNA-Flag vector (Invitrogen, Carlsbad, CA, USA). The STAT3 and JMJD2B cDNA transfections were carried out in 80% confluent cells for 72 h using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions.
Phosphorylated histone H2AX immunofluorescent staining
Both CRC cell lines were transfected with JMJD2B siRNA or negative control siRNA for 48 h, and then treated with DMSO or 50 μmol l−1 etoposide (Sigma-Aldrich, St Louis, MO, USA) for 24 h. Cells were fixed with 4% paraformaldehyde, blocked with 1% bovine serum albumin and 0.2% Triton X-100, and then incubated with an anti-phosphorylated histone H2AX (γH2AX) monoclonal antibody (Ser139, 1 : 200; Millipore, Billerica, MA, USA) and anti-H3K9me3 polyclonal antibody (1 : 300; Abcam, Cambridge, UK). Subsequently, the cells were incubated with Alexa Fluor 488 anti-mouse IgG and Alexa Fluor 594 anti-rabbit IgG (1 : 500; Molecular Probes, Eugene, OR, USA), respectively. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; SouthernBiotech, Birmingham, AL, USA). The stained cells were observed under a fluorescent microscope (LSM-710, Zeiss, Jena, Germany).
Comet assay
HCT116 and SW480 cells were transfected with JMJD2B or negative control siRNA. DNA damage was measured by the comet assay (single-cell gel electrophoresis) 48 h after transfection using the comet assay kit (Cell Biolab, San Diego, CA, USA) according to the manufacturer's protocol. Electrophoresis was run in an alkaline buffer. All of these steps were conducted under dimmed light to prevent the occurrence of additional DNA damage. After staining the DNA with ethidium bromide, individual comets were visualised using an Olympus IMT-2 inverted microscope (Olympus Center Valley, PA, USA) with epifluorescence capability under × 100 magnification. The measurement was performed using a public domain PC-image analysis programme CASP software, version 1.2.2 (CASPLab, University of Wroclaw, Wroclaw, Poland), scoring 300 comets per condition. The comet assay was repeated three times, and results are presented as percentage of DNA in tail.
Western blot analysis
Total cell proteins were extracted using RIPA lysis buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS) in the presence of a protease inhibitor cocktail (Kangcheng, Shanghai, China). For western blotting, samples were mixed with 5 × SDS–PAGE sample loading buffer (Kangcheng) and heated at 100 °C for 10 min. Proteins were resolved using SDS–PAGE and transferred to nitrocellulose filter membranes (Bio-Rad, Hercules, CA, USA). The membranes were probed using antibodies against HIF-1α (1 : 1000; BD Transduction Laboratories, Franklin Lakes, NJ, USA); JMJD2B (1 : 1000; Bethyl Laboratories, Montgomery, TX, USA); γH2AX (Ser139, 1 : 1000; Millipore); phosphorylated checkpoint kinase 2 (p-CHK2, Thr68, 1 : 1000), CHK2 (1 : 1000; Epitomics, Burlingame, CA, USA); p53(1 : 1000), p-CHK1 (Ser317, 1 : 1000), p-CHK1 (Ser345, 1 : 1000), CHK1 (1 : 1000), phosphorylated Janus kinase 2 (p-JAK2, Tyr1007/1008, 1 : 1000), JAK2 (1 : 1000), p-STAT3 (Tyr705, 1 : 1000), STAT3 (1 : 1000), and α-tubulin (1 : 1000; Cell Signaling Technology, Danvers, MA, USA). After incubation with the primary antibody overnight at 4 °C, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1 : 5000; Kangcheng) for 1 h at room temperature. Protein signals were detected by chemiluminescence (SuperSignal West Femto Maximum Sensitivity Substrate; Thermo Scientific, Rockford, IL, USA).
Cell viability assays
Cells were transfected with JMJD2B siRNA or control siRNA for 24 h after passage into 96-well plates and then incubated for 0, 6, 12, or 24 h in hypoxia. Cell viability was evaluated at different time points using the Cell Counting Kit 8 (CCK-8, Dojindo, Kumamoto, Japan) according to manufacturer's instructions in triplicate and repeated three times.
Flow cytometric analysis
Cells were harvested and fixed in 75% ethanol at −20 °C for at least 24 h, stained with propidium iodide (250 μg ml−1; Beyotime Institute of Biotechnology, Shanghai, China) containing 10 mg ml−1 RNase A (Takara, Shiga, Japan) at 37 °C for 30 min, and analysed by flow cytometry for cell cycle profiling. Apoptosis was quantified using an Annexin V Apoptosis Detection Kit I (BD Pharmingen, San Diego, CA, USA). The apoptosis and cell cycle assays were performed using a FACS Calibur flow cytometer (BD Pharmingen). The fluorescence-activated cell sorter analysis was performed in triplicate.
Senescence-associated β-galactosidase (SA-β-gal) staining
Seventy-two hours after transfection with the negative control siRNA or JMJD2B siRNA in normoxia, the cells were washed once with PBS, fixed in 4% (v/v) formaldehyde for 15 min, washed in PBS again, and incubated overnight at 37 °C in staining solution (Beyotime Institute of Biotechnology). Subsequently, the cells were washed three times with PBS before visualisation and image capture. The percentage of SA-β-gal-positive cells was determined by counting the number of blue cells under bright-field illumination, and the total number of cells in the same field under phase contrast. At least 10 fields of triplicate plates were analysed. The evaluation of senescence was made in a blinded manner.
Gene microarray analysis
HCT116 cells were treated with JMJD2B siRNA or control siRNA for 24 h, and then exposed to hypoxia for another 24 h. Total RNA was extracted from each sample using TRIzol RNA Isolation reagent (Takara) according to the manufacturer's instructions and purified with an RNeasy Mini Kit (Qiagen, Hilden, Germany). The quality and quantity of the purified RNA samples were evaluated using an Agilent 2000 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA). Data were collected with an Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix, Santa Clara, CA, USA) using standard methods and analysed using GeneSpring GX software (Agilent Technologies Inc.). Three biological replicates were tested. After normalisation, differentially expressed genes with statistical significance were identified through two compared groups (t-test, P<0.05). Relative changes in gene expression were calculated to identify genes that had been induced >2-fold and genes that had been downregulated by at least 0.5-fold after JMJD2B knockdown in hypoxia. Each up/downregulated gene was analysed for the relationship with DNA repair according to the published literature. Last, hierarchical clustering was performed to determine the extent of similarity between the up/downregulated genes.
Real-time RT–PCR analysis
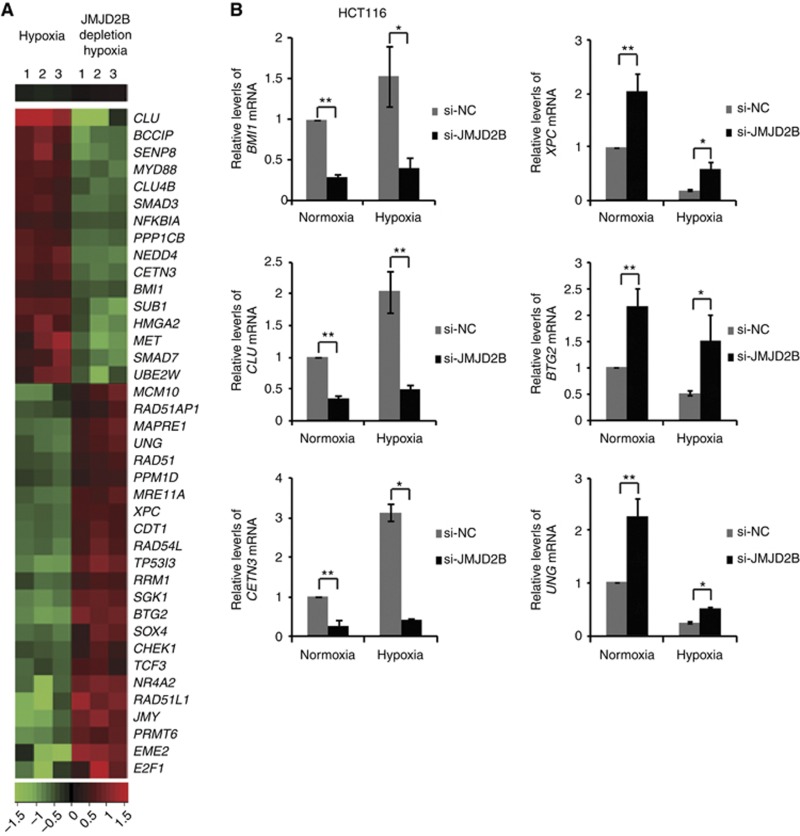
Total RNA was extracted using TRIzol RNA Isolation reagent (Takara) and reverse-transcribed using an RT Reagent Kit (Takara) according to the manufacturer's protocol. Quantitative real-time RT–PCR (qRT–PCR) was performed using SYBR Premix Ex Taq II (Takara) at 95 °C for 30 s, and 40 cycles at 95 °C for 5 s and 60 °C for 30 s using an ABI Prism 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The qRT–PCR was carried out with the primers specific for JMJD2B and selected DNA repair-related genes: BMI1, CLU, CETN3, XPC, BTG2, and UNG (Supplementary Table 1). The qRT–PCR was performed in triplicate.
In vivo experiments
HCT116 cells (1.0 × 107) were injected subcutaneously into the right flank of 4-week-old male BALB/c nude mice (Experimental Animal Centre of SIBS, Shanghai, China). After the tumours grew to 5 mm in diameter, the mice were randomly allocated (six mice per group) and treated with multipoint intratumoural injection (10 μg per 30 μl per tumour) of siRNA complexed with in vivo-jetPEI transfection reagent (Polyplus-transfection Inc., New York, NY, USA) every other day for 11 days. Tumour volume (mm3) was estimated using the formula: tumour volume (mm3)=smallest diameter2 × largest diameter/2. The tumour volume data are presented as means±s.d. The Institutional Animal Care and Use Committee approved all experimental procedures.
Immunohistochemistry staining
Immunohistochemistry was performed on tissues obtained from the mouse CRC xenograft models to examine the efficiency of JMJD2B knockdown and the γH2AX level. Antigen retrieval was performed by incubating slides in a citrate–EDTA-based solution under 1.03 × 105 Pa pressure for 2 min using a Decloaking Chamber (Biocare Medical, Walnut Creek, CA, USA). Sections were incubated overnight at 4 °C with 0.4 μg ml−1 anti-JMJD2B polyclonal rabbit antibody and anti-γH2AX monoclonal rabbit antibody (1 : 50, Ser139; Epitomics). Bound antibody was labelled with horseradish peroxidase-tagged anti-rabbit IgG (Maxim, Shanghai, China). Antibody complexes were visualised using 2,3-diaminobenzidine as a chromogen, and sections were counterstained with Mayer's haematoxylin. Last, sections of mouse CRC xenograft tissue were coverslipped and imaged.
Statistical analysis
The results are presented as means±s.d. The statistical significance of differences between groups was assessed using SPSS for Windows 11.0.1 (SPSS, Chicago, IL, USA). Statistical analyses were performed using an unpaired, two-tailed t-test, or ANOVA as appropriate. The level of significance was set at P<0.05. All analyses were performed at least three times.
Results
JMJD2B silencing induces DNA damage in both normoxia and hypoxia
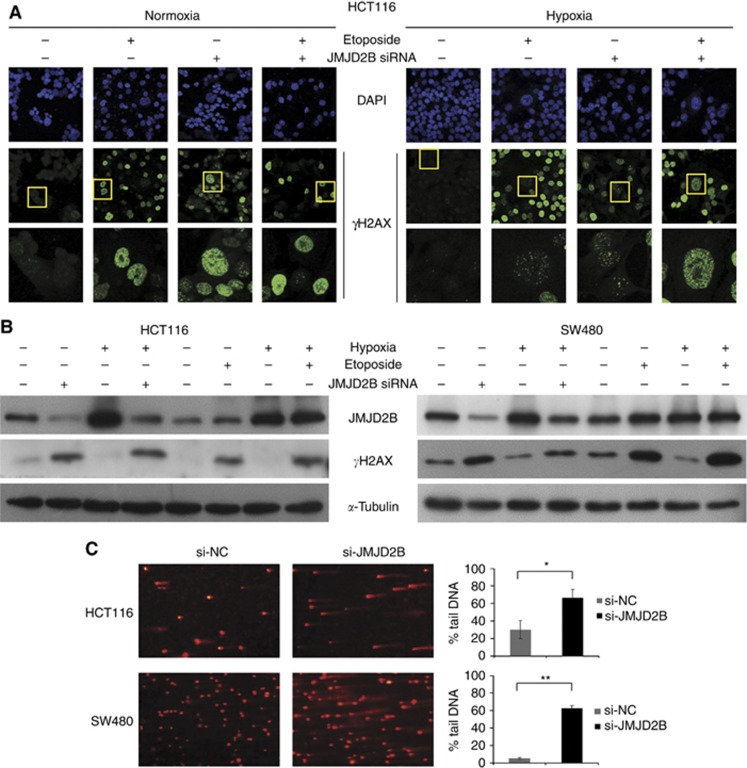
We and others have demonstrated that the histone demethylase JMJD2B functions as an oncogene by the regulation of the cell cycle, apoptosis, proliferation, and invasion in numerous tumours (Yang et al, 2010; Kawazu et al, 2011; Li et al, 2011; Shi et al, 2011; Fu et al, 2012; Kim et al, 2012). In light of stress-induced DNA damage has an essential role in the induction of cell cycle arrest, apoptosis (Harper and Elledge, 2007), and senescence (d'Adda di Fagagna, 2008), we queried whether JMJD2B has a potential role in carcinogenesis via modulation of the DDR in CRC cells. Therefore, formation of γH2AX (Ser139) nuclear foci, a characteristic of DSBs, was detected by immunofluorescence with γH2AX-specific antibodies after 48 h JMJD2B siRNA transfection. Both HCT116 and SW480 cells exposed to 50 μmol l−1 etoposide for 24 h were used as a positive control. We found that positive γH2AX staining was observed in both JMJD2B-silenced and etoposide-treated cells (Figure 1A and Supplementary Figure 2A, second pannel). In higher-magnification images, numerous γH2AX-positive foci could be observed clearly (Figure 1A and Supplementary Figure 2A, third panel); however, H3K9me3 did not colocalise with the γH2AX foci (data not shown). Consistently, western blotting revealed remarkable increase of γH2AX expression when JMJD2B was knocked down by siRNA in HCT116 and SW480 cells not only in normoxia but also in hypoxia, similar to that of etoposide treatment (Figure 1B). To directly assess the effect of JMJD2B silencing on DNA damage, comet assay was applied after transfection with JMJD2B siRNA in HCT116 and SW480 cells for 48 h. Depletion of JMJD2B resulted in DNA damage, as monitored by formation of a comet-like tail following single-cell gel electrophoresis (Figure 1C, left). Thereafter, DNA damage was measured by the percentage of DNA in tail. There are significant increases in percentage of DNA in tail after JMJD2B siRNA transfection (Figure 1C, right). From these results, it is evident that JMJD2B silencing could induce DNA damage in HCT116 and SW480 cells in both normoxia and hypoxia.
Figure 1.
JMJD2B silencing induced DNA damage in both normoxia and hypoxia. (A) Knockdown of JMJD2B causes γH2AX (Ser139) nuclear foci formation under normoxia and hypoxia. Representative immunofluorescence staining for γH2AX (Ser139) in HCT116 cells under normoxia (left) and hypoxia (right). Etoposide was applied as a positive control. (B) Effect of JMJD2B on γH2AX (Ser139) levels. Etoposide was applied as a positive control. Representative western blot analysis (left to right) from normoxia negative control siRNA transfected, normoxia JMJD2B siRNA transfected, hypoxia negative control siRNA transfected, hypoxia JMJD2B siRNA transfected, and normoxia DMSO treated, normoxia etoposide treated, hypoxia DMSO treated, hypoxia etoposide treated HCT116 (left) and SW480 (right) cells. (C) JMJD2B silencing induced DNA damage. Comet assay was applied after 48 h of JMJD2B siRNA transfection in HCT116 and SW480 cells under normoxia. Representative results of three independent experiments are shown (left). The extent of DNA damage was determined by measuring the percentage of DNA in tail for 300 individual comets. Data were quantified and graphed (right). HCT116 *P<0.05, SW480 **P<0.01 vs Si-NC. Abbreviations: si-NC=the negative control siRNA; si-JMJD2B=the siRNA-targeted JMJD2B.
HIF-1α silencing induces DNA damage partially through JMJD2B inactivation in hypoxia
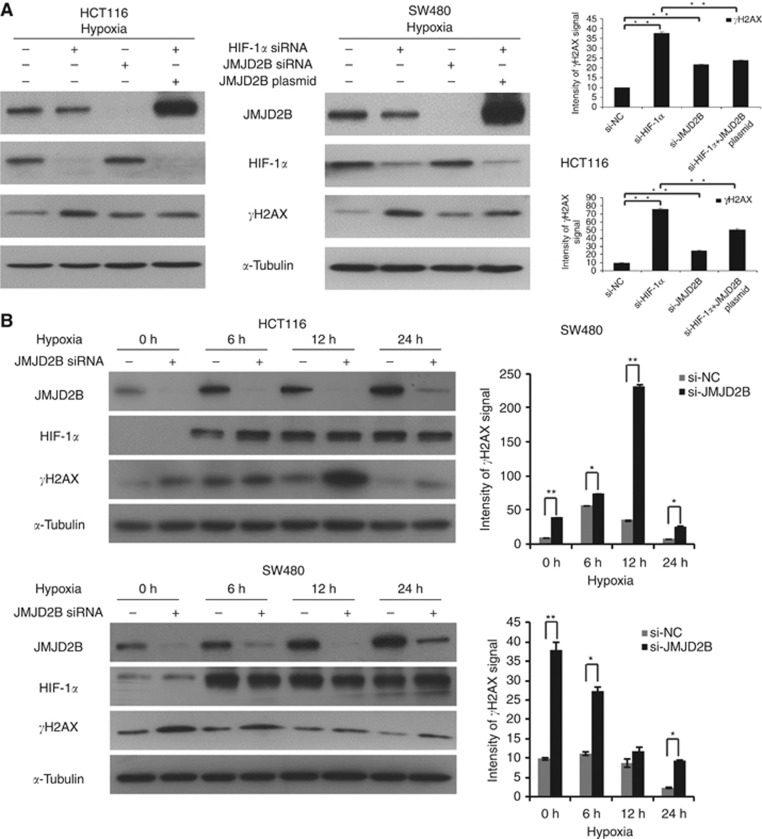
We and others have shown that JMJD2B can be upregulated in hypoxia in a HIF-1α-dependent manner (Beyer et al, 2008; Pollard et al, 2008; Fu et al, 2012); moreover, hypoxia can regulate the DDR, raising the question of whether JMJD2B is still required for DDR mediated by HIF-1α in hypoxia. To address this question, after transfection with HIF-1α siRNA for 48 h, CRC cells were then exposed to hypoxia for 24 h. In both cell lines, HIF-1α suppression not only reduced JMJD2B expression, but also significantly activated the DDR (P<0.05; Figure 2A). As expected, a marked decrease in the phosphorylation of H2AX was determined to correspond to the ectopic expression of JMJD2B in HIF-1α-depleted CRC cells (Figure 2A). Taken together, these results clearly indicate that in hypoxia, HIF-1α can partially mediate the DDR by regulating JMJD2B expression in CRC cells.
Figure 2.
HIF-1α silencing induces DNA damage in a JMJD2B-dependent manner. (A) Representative western blot analysis from negative control siRNA, HIF-1α siRNA, JMJD2B siRNA, or HIF-1α siRNA and JMJD2B plasmid-transfected HCT116 and SW480 cells (left). Quantification of γH2AX (Ser139) levels (right). Both HIF-1α and JMJD2B siRNA transfection resulted in a significant increase in the level of γH2AX (Ser139; **P<0.01 vs negative control siRNA). Ectopic expression of JMJD2B in HIF-1α-depleted CRC cells largely alleviated HIF-1α silencing-induced γH2AX (Ser139) level (**P<0.01 vs HIF-1α siRNA). Band intensities were measured using ImageJ, normalised to α-tubulin, and presented as percentages relative to the levels of γH2AX (Ser139) with negative control siRNA. All data from at least three independent experiments are presented as mean±s.d. (B) Knockdown of JMJD2B increased γH2AX expression at different time points in hypoxia. Representative western blot analysis from negative control or JMJD2B siRNA-transfected HCT116 (upper left) and SW480 (lower left) cells for 0, 6, 12, or 24 h hypoxia. Quantification of γH2AX (Ser139) levels at indicated times (right).
JMJD2B silencing induces DNA damage via the ataxia telangiectasia-mutated (ATM) and ATM- and Rad3-related (ATR) pathways
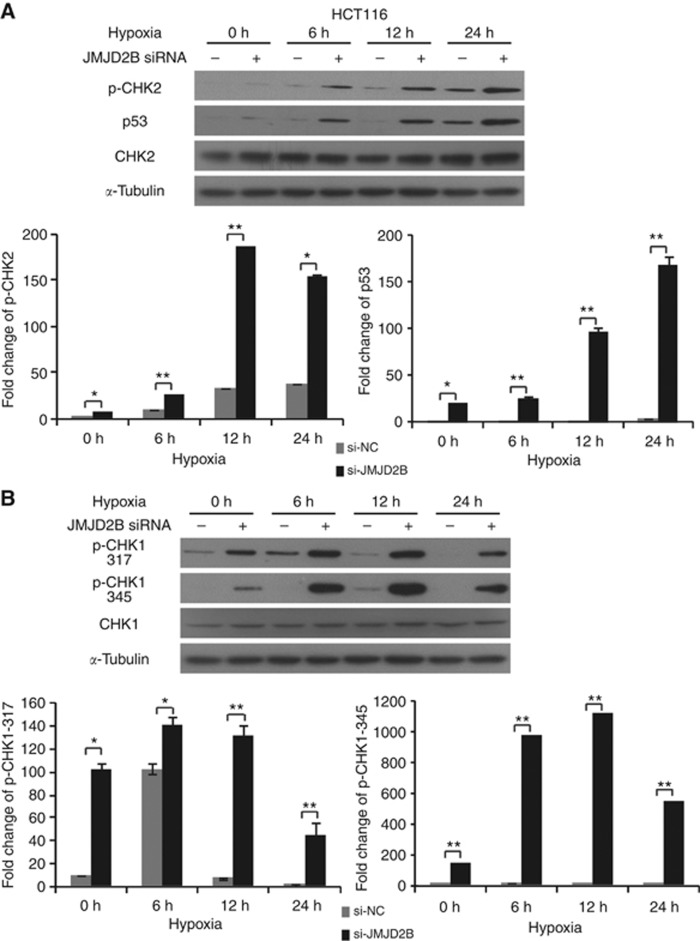
DNA damage, especially in the form of DSBs, results in the activation of a complex signalling network characterised by the activation of ATM and ATR kinase. Two structurally unrelated but functionally similar protein serine/threonine kinases Chk2 and Chk1 are the substrates of ATM and ATR. Chk2 is phosphorylated by ATM protein at Thr68 and thus activated (Ahh et al, 2000), whereas Chk1 activation requires phosphorylation at Ser317 and Ser345 in an ATR-dependent manner (Zhao and Piwnica-Worms, 2001). Thus, to further identify the DDR kinases induced by JMJD2B silencing, we performed a time-course study to define the role of ATM and ATR pathways. CRC cells were pretreated with JMJD2B siRNA for 24 h, and then incubated in hypoxia for 0, 6, 12, or 24 h. Six-hour exposure to hypoxia was sufficient to stimulate JMJD2B; in parallel, γH2AX levels began to increase in JMJD2B-silenced cells as early as 6 h after hypoxia treatment, and reached maximum levels at 12 h in the HCT116 cells, whereas γH2AX expression in JMJD2B-silenced SW480 cells was relatively lower in hypoxia than normoxia (Figure 2B). In JMJD2B siRNA-treated HCT116 cells, p-Chk2 was detected as early as 6 h and their expression was sustained for 24 h in hypoxia (Figure 3A). Furthermore, Chk1 was also activated (Figure 3B), indicating JMJD2B silencing significantly activated the ATM and ATR pathways in HCT116 cells. The unexpected was that in SW480 cells, only Chk1 was phosphorylated in hypoxia after JMJD2B knockdown (Supplementary Figure 2), almost corresponding to the γH2AX levels. It was documented that the activation of ATM and ATR leads to the phosphorylation of histone H2AX, which has an important role in the DDR associated with the stimulation of DNA repair, activation of cell cycle checkpoint, and eventually induction of apoptosis (Harper and Elledge, 2007). Therefore, the data suggest that the ATM and ATR pathways may be involved in the regulation of JMJD2B silencing-induced DDR.
Figure 3.
JMJD2B silencing induces DNA double-strand breaks via the ATM-CHK2 and/or ATR-CHK1 pathway. (A) JMJD2B silencing upregulated p-CHK2-p53 expression. Representative western blot analysis from negative control or JMJD2B siRNA-transfected HCT116 cells at different time points (upper). Quantification of p-CHK2 (lower left) and p53 (lower right) levels (*P<0.05, **P<0.01 vs Si-NC). (B) Downregulation of JMJD2B markedly elevated p-CHK1 (Ser317/345) protein levels. Representative western blot analysis from negative control or JMJD2B siRNA-transfected HCT116 cells at indicated times (upper). Quantification of p-CHK1 (Ser317; lower left) and p-CHK1 (Ser345; lower right) levels. Band intensities were measured using ImageJ, normalised to α-tubulin, and presented as percentages relative to the levels of indicated protein with negative control siRNA at indicated times, respectively (*P<0.05, **P<0.01 vs Si-NC). All data from at least three independent experiments are presented as mean±s.d.
JMJD2B silencing-induced DNA damage mediates cell cycle arrest, apoptosis, and senescence
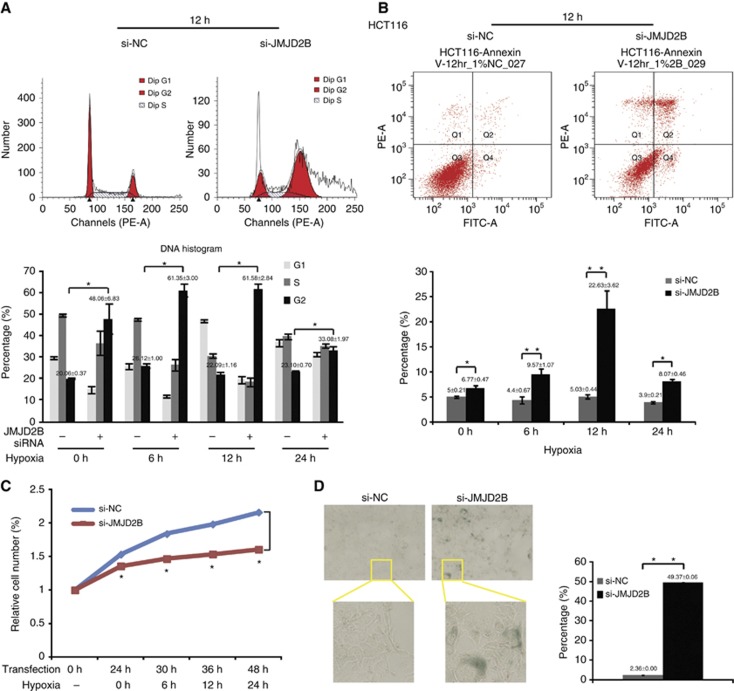
To investigate the role of JMJD2B in the regulation of cancer cell survival and senescence, we examined the growth profiles of JMJD2B-silenced HCT116 and SW480 cells in a time-course study in hypoxia. Compared with the control group, there was a significant increase of HCT116 cells with maximum 4N content at 12 h, and an increase of SW480 cells with maximum 2N content at 24 h after JMJD2B silencing in hypoxia, indicative of G2 and G1 phase arrest, respectively (Figure 4A and Supplementary Figure 3A). Besides, an increase number of cells displaying >4N DNA content was observed in JMJD2B-depleted HCT116 cells, whereas not in the control cells and SW480. Furthermore, as shown in Figure 4C and Supplementary Figure 3C, JMJD2B silencing remarkably reduced the growth of HCT116 and SW480 cells in hypoxia as measured by CCK-8 assay (P<0.05). This effect was accompanied by a similar significant accumulation of apoptotic cells after JMJD2B silencing (P<0.05; Figure 4B and Supplementary Figure 3B). Cellular senescence is an anti-proliferative programme that functions as a potent tumour-suppressor mechanism (Collado and Serrano, 2011). Morphologically, JMJD2B-silenced cells were larger and appeared flattened (data not shown), which is a feature of senescence (Itahana et al, 2007). To further address whether JMJD2B silencing induced senescence, we carried out SA-β-gal staining. Positive SA-β-gal staining was observed at 72 h post-transfection in about 49% and 20% of JMJD2B-silenced HCT116 and SW480 cells, respectively, but only <2% of the control group were SA-β-gal–positive (P<0.05; Figure 4D and Supplementary Figure 3D). Overall, our results indicated that JMJD2B silencing-induced DDR has an important role in the induction of cell cycle arrest, apoptosis, and senescence.
Figure 4.
JMJD2B is essential for CRC cell survival and senescence. (A) Knockdown of JMJD2B impaired cell cycle transition of HCT116 cells (upper). G2-M phase arrest was detected after 24 h of JMJD2B siRNA transfection. Data were quantified and graphed at indicated times (lower) (*P<0.05 vs Si-NC). (B) Knockdown of JMJD2B caused significant apoptosis in HCT116 cells in 0, 6, 12, and 24 h hypoxia (*P<0.05, **P<0.01 vs Si-NC). (C) Effect of JMJD2B on HCT116 cell viability measured by CCK-8 assay after 0, 6, 12, and 24 h of incubation under hypoxia. Transfection with JMJD2B siRNA (si-JMJD2B) induced significant decrease in cell viability (red line). Each time point is represented as percentage relative to 0 h at transfection. Data show the mean percentage±s.d. of three independent experiments (*P<0.05 vs Si-NC). (D) Senescence was significantly induced in HCT116 cells by JMJD2B silencing via SA-β-gal staining assay. Senescent cells were stained blue (left). Senescent cells were counted in 10 randomised fields of vision, and results are presented as percentage of SA-β-gal-positive cells in total cells in the graph (right). **P<0.01 vs Si-NC. Abbreviations: si-NC=the negative control siRNA; si-JMJD2B=the siRNA-targeted JMJD2B. The full colour version of this figure is available at British Journal of Cancer online.
Alterations in DNA damage repair gene expression are involved in JMJD2B suppression-induced DNA damage
In order to probe the DNA repair genes regulated by JMJD2B, we carried out a gene expression profiling study. HCT116 cells were transfected with negative control siRNA or JMJD2B siRNA and then exposed to 1% hypoxia for 24 h, and gene expression was analysed using the Affymetrix Human Genome U133 Plus 2.0 Array. We identified 16 downregulated genes (criteria: fold change <0.5; P<0.05, t-test) and 23 upregulated genes (criteria: fold change >2; P<0.05, t-test) involved in DNA repair after JMJD2B suppression (Figure 5A). Functional analysis of these genes indicated that they were related to a number of key DNA repair processes, including regulation of homologous recombination repair (MET, RAD51AP1, RAD54L, RAD51, CHEK1, SMAD3), non-homologous end-joining repair (HMGA2), nucleotide excision repair (XPC, PPM1D, CETN3), base excision repair (PPM1D), and DSB repair (MRE11A, HMGA2, BMI1, NR4A2). Validations of three selected genes whose expression was JMJD2B dependent (BMI1, CETN3, CLU) from the 16 downregulated genes and three from the 23 upregulated genes (XPC, UNG, BTG2) were confirmed with qRT–PCR (Figure 5B and Supplementary Figures 4A and B). Overall, these data imply that JMJD2B may have an important role in regulating DNA repair.
Figure 5.
JMJD2B regulates DNA repair gene expression in HCT116 cells. (A) Heatmap of JMJD2B-regulated DNA damage repair genes in hypoxia by Affymetrix gene microarray with triplicates. Sixteen genes were identified downregulated <0.5-fold and 23 genes upregulated >2-fold with JMJD2B knockdown. (B) qRT–PCR validation of the expression of JMJD2B-regulated DNA damage repair genes. Data were normalised to 18s ribosomal RNA, and presented as fold changes relative to the mRNA levels of indicated genes in normoxia. Three replicates of qRT–PCR were performed, and repeated at least three times. BMI1, CLU, and CETN3 mRNA levels were significantly downregulated with JMJD2B siRNA transfection. XPC, BTG2, and UNG mRNA levels were markedly upregulated with JMJD2B silencing. *P<0.05, **P<0.01 vs si-NC. Abbreviations: si-NC=the negative control siRNA; si-JMJD2B=the siRNA-targeted JMJD2B.
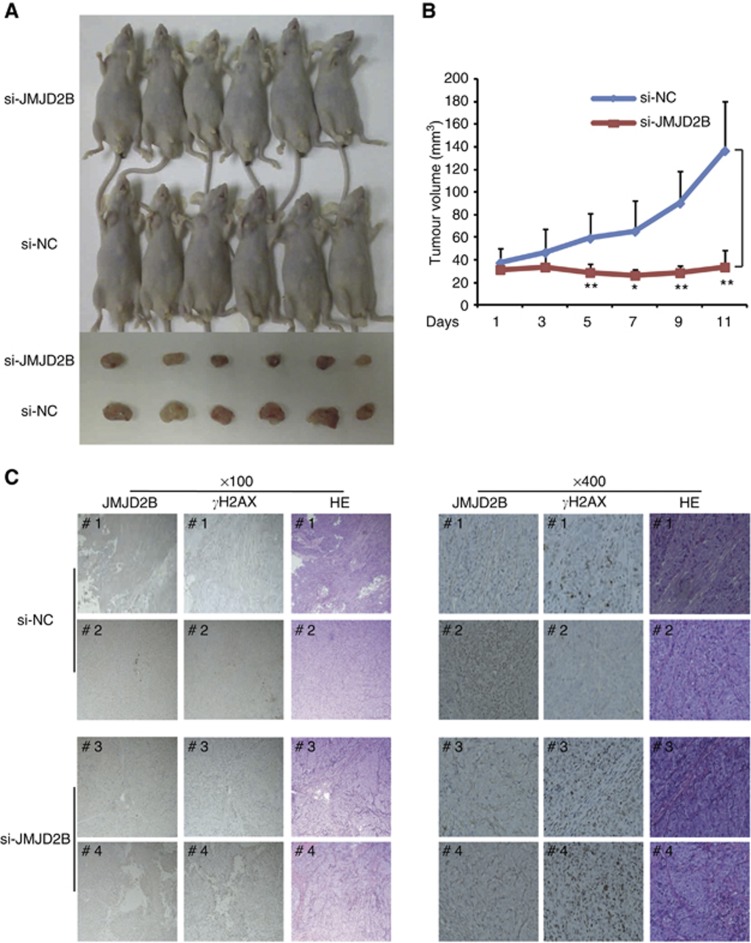
JMJD2B inhibition suppressed tumour growth in the CRC xenograft model
To further confirm the impact of JMJD2B on in vivo tumour growth and DDR, we generated a mouse model and evaluated the effect of targeting JMJD2B with siRNAs. As shown in Figures 6A and B, the tumour volume of the JMJD2B siRNA group was significantly smaller than that of the control siRNA group (P<0.05). Furthermore, we examined JMJD2B and γH2AX expression in the xenograft tissues by immunohistochemistry. We confirmed that the intratumoural injection of JMJD2B-targeting siRNA knocked down JMJD2B expression (Figure 6C). Consistent with the observations made in HCT116 and SW480 cells, JMJD2B knockdown led to γH2AX accumulation in the treated tumour tissues. Taken together, these data strongly support our notion that targeting JMJD2B could exert anti-cancer effects in vivo on CRC, probably by the activation of the DDR.
Figure 6.
JMJD2B silencing in vivo significantly suppressed CRC xenografts growth and led to γH2AX accumulation. (A) Effect of JMJD2B depletion on the tumour volumes of xenografts in nude mice. Abbreviations: si-NC, the negative control siRNA; si-JMJD2B, the siRNA-targeted JMJD2B. (B) Loss of JMJD2B reduces tumour growth rate. A total of 1.0 × 107 HCT116 cells were injected into the right flank of BALB/c nude mice. After the tumours grew to 5 mm in diameter, a negative control or JMJD2B siRNA mixed with in vivo-jetPEI transfection reagent, respectively, were injected intratumourally every other day for 11 days. Tumour volumes were measured at the indicated intervals. The tumour volume data are presented as means±s.d. *P<0.05, **P<0.01 vs si-NC. (C) JMJD2B knockdown led to γH2AX accumulation. Representative immunohistochemistry of JMJD2B and γH2AX in negative control (upper) and JMJD2B silencing (lower) tumour tissues. Abbreviation: HE=haematoxylin and eosin staining.
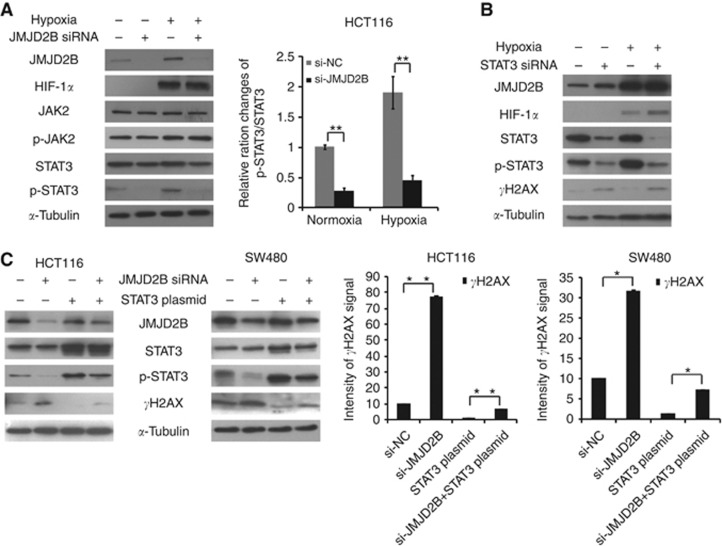
JMJD2B silencing induces DNA damage by the suppression of STAT3 signalling
Cancer-related signalling pathways are critical to carcinogenesis. As our previous studies (Xiong et al, 2008; Du et al, 2012) indicated that the inhibition of JAK2/STAT3 signalling can induce CRC cell apoptosis and cell cycle arrest, we speculated that JAK2/STAT3 may be involved in the JMJD2B silencing–induced DNA damage. As seen in Figure 7A and Supplementary Figure 5A, although a slight decrease of total STAT3 was visually observed, p-STAT3 expression significantly decreased in JMJD2B-silenced cells in both normoxia and hypoxia, which was confirmed upon quantification. In addition, no change was seen at the protein level for p-JAK2, indicating that JMJD2B were working, in part, through modulation of STAT3 phosphorylation. To further demonstrate whether STAT3 is involved in JMJD2B silencing-induced DNA damage, we suppressed STAT3 expression via siRNA for 48 h, and detected the phosphorylation level of H2AX in HCT116 and SW480 cells. Compared with the controls, STAT3 knockdown resulted in increased H2AX phosphorylation at Ser139 (Figure 7B and Supplementary Figure 5B). In addition, transduction of STAT3 cDNA into the JMJD2B-depleted cells abolished H2AX phosphorylation induced by JMJD2B suppression (Figure 7C). Taken together, our results clearly show that JMJD2B silencing induced DNA damage in HCT116 and SW480 cells partially by the regulation of the STAT3 signalling pathway, which contributed to the induction of cell cycle arrest, apoptosis, and senescence.
Figure 7.
JMJD2B silencing induces DNA damage by the suppression of STAT3 signalling. (A) Effect of JMJD2B on JAK2/STAT3 signal pathway. Representative western blot analysis from negative control, JMJD2B siRNA-transfected HCT116 cells in normoxia and hypoxia (left). Quantification of pSTAT3/STAT3 ratios (right). JMJD2B siRNA transfection resulted in a marked decrease in the ratios of pSTAT3/STAT3 during both normoxia and hypoxia. Band intensities were measured using ImageJ, normalised to α-tubulin, and presented as percentages relative to the pSTAT3/STAT3 ratios with negative control siRNA. **P<0.01 vs negative control siRNA. Abbreviations: si-NC, the negative control siRNA; si-JMJD2B, the siRNA-targeted JMJD2B. (B) STAT3 silencing induced H2AX phosphorylation. Representative western blot analysis from negative control, STAT3 siRNA-transfected HCT116 cells in normoxia and hypoxia. (C) Knockdown of JMJD2B caused DSBs through STAT3 pathway. Representative western blot analysis from negative control, JMJD2B siRNA, STAT3 plasmids or JMJD2B siRNA and STAT3 plasmid-transfected HCT116 and SW480 cells (left). Quantification of γH2AX (Ser139) levels (right). Ectopic expression of STAT3 in JMJD2B-depleted CRC cells largely alleviated JMJD2B silencing-induced γH2AX (Ser139) levels. Band intensities were measured using ImageJ, normalised to α-tubulin, and presented as percentages relative to the levels of γH2AX (Ser139) with negative control siRNA. *P<0.05, **P<0.01. All data from at least three independent experiments are presented as mean±s.d.
Discussion
Abnormalities of the histone demethylase JMJD2B are involved in the pathogenesis of several cancers, including CRC. However, the molecular mechanism by which dysregulated JMJD2B contributes to the malignant phenotype of CRC has not been fully clarified. In the present study, we examined the roles and mechanisms of JMJD2B-mediated DDR in human CRC (HCT116 and SW480) cell culture models in vitro and in a CRC xenograft model in vivo.
Using immunofluorescence and western blotting, we confirmed that JMJD2B knockdown increased the γH2AX level in the CRC cell lines in both normoxia and hypoxia. Consistently, the γH2AX level in the available tumour tissues harvested from the xenograft model was markedly increased in the JMJD2B-silenced tumours. Two distinct kinase signalling cascades, the ATM-CHK2 and ATR-CHK1 pathways, which are activated by DSBs and single-stranded DNA, respectively, primarily orchestrate cellular responses to DNA damage. The activation of ATM and ATR leads to the phosphorylation of H2AX (Ser139). Our data show that JMJD2B silencing activated both the ATM-CHK2 and ATR-CHK1 pathways in HCT116 cells, whereas it activated only the ATR-CHK1 pathway in SW480 cells, which may reflect cell type-specific responses. Moreover, the decrease of DNA repair factor functions also induces similar checkpoint activation, senescence, and finally, DNA damage because of the accumulation of unrepaired DNA lesions. Recently, it was reported that p53 regulation of JMJD2B facilitates the repair of damaged heterochromatin DNA (Zheng et al, 2013). In addition, Leah et al demonstrated that overexpression of Kdm4b enhanced the repair of DSBs, which is dependent on its catalytic domain as H3K9me3 demethylase (Young et al, 2013). These seem that regulation of histone methylation may have a vital role in the maintenance of genomic stability, of which the repair of DNA damage is an important component. Interestingly, we found the decreased expression of 16 DNA repair-related genes following JMJD2B suppression, consistent with the known ability of JMJD2B to specifically remove repressive H3K9me3 markers on target promoters (Fodor et al, 2006; Shi et al, 2011). Thus, as a histone demethylase, JMJD2B may link DDR-associated chromatin changes to localised transcriptional repression near the sites of DNA damage. Taken together, we have demonstrated that JMJD2B regulates multiple processes in the DDR based on the following findings: first, H2AX phosphorylation, a hallmark of DDR, was induced in CRC cell lines in both normoxia and hypoxia after JMJD2B suppression. Second, JMJD2B regulated the DDR of CRC cells by activating ATM and ATR signalling. Third, JMJD2B silencing downregulated the genes involved in DSB repair.
As the DDR network is tied to cellular processes such as cell cycle arrest, senescence, and apoptosis (Harper and Elledge, 2007; d'Adda di Fagagna, 2008), we further evaluated the biological significance of JMJD2B silencing-induced DDR in the pathogenesis of CRC cells. Our results indicated that JMJD2B downregulation was associated with a significant induction of apoptosis and senescence, and cell cycle arrest in the G1 or G2 phase. More interestingly, we found JMJD2B suppression in HCT116 increased the percentage of cells with a DNA content greater than 4N associated with aneuploidy (Figure 4A), whereas not in SW480, probably due to a cell line-dependent difference. As aneuploidy is a consequence of chromosome instability, which can be considered as one form of genomic instability (Draviam et al, 2004), our data may further confirm that JMJD2B depletion increased genomic instability in HCT116. Senescence is a cellular response preventing tumourigenesis. Recent reports have proposed an important role for demethylase in senescence. JMJD2A depletion triggered cellular senescence by negatively regulating the p53 pathway with its catalytic activity (Mallette and Richard, 2012) and KDM2B knockdown resulted in premature senescence via modulating let-7b-EZH2 and p15Ink4b (Tzatsos et al, 2011; He et al, 2008). Moreover, JARID1 contributed to senescence by facilitating the silencing of some retinoblastoma gene targets (Chicas et al, 2012), whereas JMJD3 through the activation of the INK4A-ARF locus (Agger et al, 2009). Because senescence, initiated by oncogene activation, telomere dysfunction, or other stimulus, is frequently associated with DNA damage (Di Micco et al, 2006), our results, therefore, strongly suggest that JMJD2B silencing-induced senescence could result from cellular response to DNA damage. Consistent with the in vitro observation, we demonstrated that in xenograft mouse model, suppression of JMJD2B in vivo also significantly delayed tumour growth. Thus, our in vivo and in vitro data both further imply that JMJD2B is a potential therapeutic target in CRC.
Owing to an inadequate and irregular blood supply, the majority of solid tumours contain hypoxic regions. Previous studies have indicated that hypoxia is associated with genetic instability, subsequently increased DNA damage, enhanced mutagenesis, and functional impairment of the DNA repair pathways, often occurs in cells exposed to prolonged hypoxia (Bindra et al, 2005; Bindra and Glazer, 2007b) and severe hypoxia (Pires et al, 2010). Jumonji domain-containing protein 2B is a direct target of HIF-1α, and is regulated by HIF-1α. Our data suggest that HIF-1α knockdown induced the DDR in cells exposed to 1% hypoxia for 24 h, accompanied by JMJD2B downregulation; furthermore, this change coincided with the suppression of JMJD2B (Figure 2A). It has also been reported that HIF-1 could restrain etoposide-induced DNA damage in hypoxia (Sullivan and Graham, 2009). Thus, it is very likely that HIF-1α may partially regulate the DDR through JMJD2B.
Previously, our laboratory discovered that inhibition of JAK2/STAT3 signalling induced CRC cell apoptosis through the mitochondrial pathway, accompanied by the activation of the caspase cascade and increased levels of reactive oxygen species (ROS; Du et al, 2012). Reactive oxygen species are one of the common endogenous DNA damage stresses (Vilenchik and Knudson, 2003), whereas STAT3 is constitutively present in the mitochondria (Wegrzyn et al, 2009; Boengler et al, 2010), which is one of the major sources of ROS in the cell (Turrens, 2003). Increasing evidence has indicated that STAT3 silencing induces more ROS (Dixit et al, 2009; Tierney et al, 2012) and/or disables the function of scavenging ROS generation (Negoro et al, 2001), which modulates susceptibility to chemotherapeutic agents (Alas and Bonavida, 2003; Dixit et al, 2009; Kulesza et al, 2013) through increased apoptosis and DNA damage (Bonner et al, 2011). Specifically, the microarray data showed some JMJD2B-regulated genes involved in DNA repair are also linked to the STAT3 pathway (Figure 5). MyD88 has a key role in mediating STAT3 phosphorylation (Yamawaki et al, 2010) and STAT3 might regulate BMI1 gene through SALL4 (Bard et al, 2009). Our results reveal that JMJD2B silencing downregulated STAT3 phosphorylation; thus, we speculated that STAT3 signalling might serve as a bridge linking JMJD2B and DDR. This notion is supported by our observation that STAT3 suppression induced γH2AX accumulation, whereas STAT3 overexpression mitigated endogenous DNA damage in CRC cells. Although several models have been suggested, the accurate mechanisms of how STAT3 regulates the DDR remain elusive.
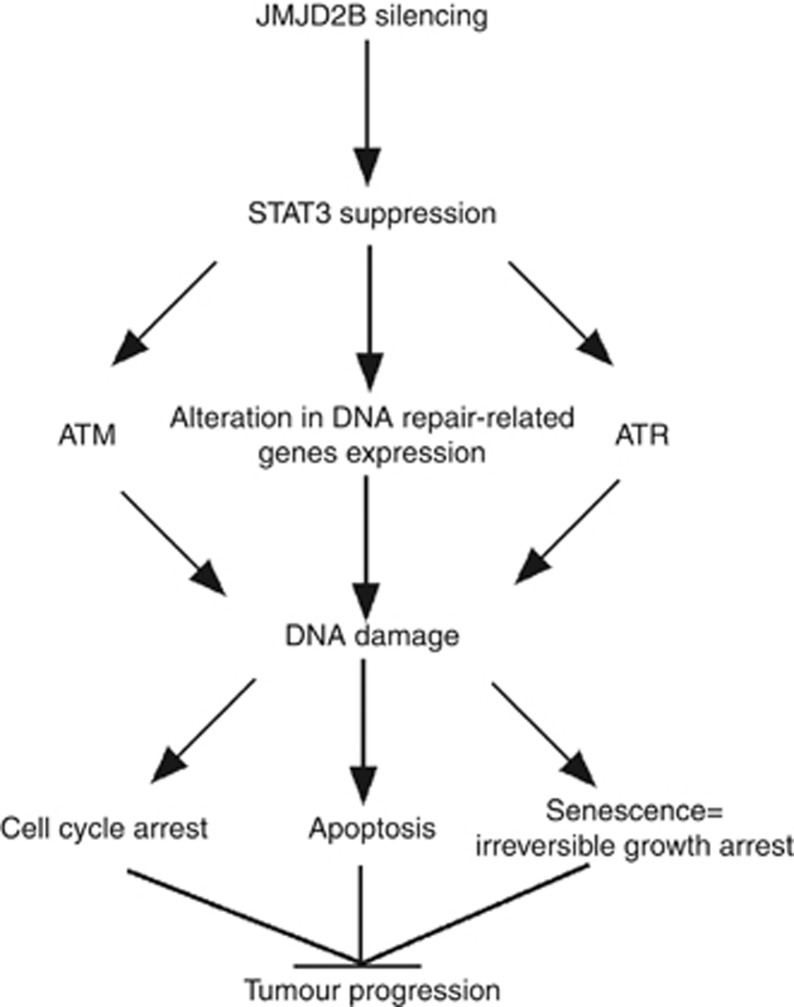
In conclusion, the major finding of this study is that JMJD2B silencing-induced DNA damage triggers senescence, apoptosis, and cell cycle arrest in HCT116 and SW480 cells by the suppression of STAT3 signalling in both normoxia and hypoxia. As commonly used anti-cancer agents exert their cytotoxic effects by inducing DNA damage through diverse mechanisms (Lord and Ashworth, 2012), deficient DNA damage signalling pathways and DNA repair were thought to be a key determinant of chemoresistance and radioresistance. Our results, therefore, first identify a novel link between the JMJD2B–STAT3 pathway and the DDR in both normoxia and hypoxia. Overall, the current study provides novel insights into the signalling networks involved in the possible carcinogenic effects of the JMJD2B gene (summarised in Figure 8), which provide further support for targeting JMJD2B in the potential treatment of human CRC.
Figure 8.
Model for JMJD2B silencing on tumour suppression through DNA damage. JMJD2B silencing induced DNA damage through ATM, ATR pathway, and regulating DNA damage repair genes, which is in a STAT3-dependent manner. Enhanced DNA damage resulted in cell cycle arrest, apoptosis and senescence, and suppressing tumour progression.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81172321, to C.Y.X), and A3 Foresight Program from Natural Science Foundation of China (No. 31261140372).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahh JY, Schwarz JK, Piwnica-Worms H, Canman CE. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 2000;60:5934–5936. [PubMed] [Google Scholar]
- Alas S, Bonavida B. Inhibition of constitutive STAT3 activity sensitizes resistant non-Hodgkin's lymphoma and multiple myeloma to chemotherapeutic drug-mediated apoptosis. Clin Cancer Res. 2003;9:316–326. [PubMed] [Google Scholar]
- Bard JD, Gelebart P, Amin HM, Young LC, Ma Y, Lai R. Signal transducer and activator of transcription 3 is a transcriptional factor regulating the gene expression of SALL4. FASEB J. 2009;23:1405–1414. doi: 10.1096/fj.08-117721. [DOI] [PubMed] [Google Scholar]
- Beyer S, Kristensen MM, Jensen KS, Johansen JV, Staller P. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J Biol Chem. 2008;283:36542–36552. doi: 10.1074/jbc.M804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M, Pierce AJ, Bristow RG, Classon MK, Glazer PM. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 2005;65:11597–11604. doi: 10.1158/0008-5472.CAN-05-2119. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. Co-repression of mismatch repair gene expression by hypoxia in cancer cells: role of the Myc/Max network. Cancer Lett. 2007;252:93–103. doi: 10.1016/j.canlet.2006.12.011. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene. 2007;26:2048–2057. doi: 10.1038/sj.onc.1210001. [DOI] [PubMed] [Google Scholar]
- Boengler K, Hilfiker-Kleiner D, Heusch G, Schulz R. Inhibition of permeability transition pore opening by mitochondrial STAT3 and its role in myocardial ischemia/reperfusion. Basic Res Cardiol. 2010;105:771–785. doi: 10.1007/s00395-010-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JA, Yang ES, Trummell HQ, Nowsheen S, Willey CD, Raisch KP. Inhibition of STAT-3 results in greater cetuximab sensitivity in head and neck squamous cell carcinoma. Radiother Oncol. 2011;99:339–343. doi: 10.1016/j.radonc.2011.05.070. [DOI] [PubMed] [Google Scholar]
- Chan N, Koritzinsky M, Zhao H, Bindra R, Glazer PM, Powell S, Belmaaza A, Wouters B, Bristow RG. Chronic hypoxia decreases synthesis of homologous recombination proteins to offset chemoresistance and radioresistance. Cancer Res. 2008;68:605–614. doi: 10.1158/0008-5472.CAN-07-5472. [DOI] [PubMed] [Google Scholar]
- Chicas A, Kapoor A, Wang X, Aksoy O, Evertts AG, Zhang MQ, Garcia BA, Bernstein E, Lowe SW. H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence. Proc Natl Acad Sci USA. 2012;109:8971–8976. doi: 10.1073/pnas.1119836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Serrano M. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer. 2011;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre' M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d'Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Dixit D, Sharma V, Ghosh S, Koul N, Mishra PK, Sen E. Manumycin inhibits STAT3, telomerase activity, and growth of glioma cells by elevating intracellular reactive oxygen species generation. Eree Radic Biol Med. 2009;47:364–374. doi: 10.1016/j.freeradbiomed.2009.04.031. [DOI] [PubMed] [Google Scholar]
- Draviam VM, Xie S, Sorger PK. Chromosome segregation and genomic stability. Curr Opin Genet Dev. 2004;14:120–125. doi: 10.1016/j.gde.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Du W, Hong J, Wang YC, Zhang YJ, Wang P, Su WY, Lin YW, Lu R, Zou WP, Xiong H, Fang JY. Inhibiton of JAK2/STAT3 signalling induces colorectal cancer cell apoptosis via mitochondrial pathway. J Cell Mol Med. 2012;16:1878–1888. doi: 10.1111/j.1582-4934.2011.01483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor BD, Kubicek S, Yonezawa M, O'Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Chen L, Yang J, Ye T, Chen Y, Fang J. HIF-1α-induced histone demethylase JMJD2B contributes to the malignant phenotype of colorectal cancer cells via an epigenetic mechanism. Carcinogenesis. 2012;33:1664–1673. doi: 10.1093/carcin/bgs217. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat Struct Mol Biol. 2008;15:1169–1175. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Campisi J, Dimri GP. Methods to detect biomarkers of cellular senescence: the senescence-associated beta-galactosidase assay. Methods Mol Biol. 2007;371:21–31. doi: 10.1007/978-1-59745-361-5_3. [DOI] [PubMed] [Google Scholar]
- Kawazu M, Saso K, Tong KI, McQuire T, Goto K, Son DO, Wakeham A, Miyagishi M, Mak TW, Okada H. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One. 2011;6:e17830. doi: 10.1371/journal.pone.0017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JG, Yi JM, Park SJ, Kim JS, Son TG, Yang K, Yoo MA, Heo K. Histone demethylase JMJD2B-mediated cell proliferation regulated by hypoxia and radiation in gastric cancer cells. Biochim Biophys Acta. 2012;1819:1200–1207. doi: 10.1016/j.bbagrm.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Kulesza DW, Carré T, Chouaib S, Kaminska B. Silencing of the transcription factor STAT3 sensitizes lung cancer cells to DNA damaging drugs, but not to TNFα- and NK cytotoxicity. Exp Cell Res. 2013;19:506–516. doi: 10.1016/j.yexcr.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Li W, Zhao L, Zang W, Liu Z, Chen L, Liu T, Xu D, Jia J. Histone demethylase JMJD2B is required for tumor cell proliferation and survival and is overexpressed in gastric cancer. Biochem Biophys Res Commun. 2011;416:372–378. doi: 10.1016/j.bbrc.2011.11.045. [DOI] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- Mallette FA, Mattiroli F, Cui G, Young LC, Hendzel MJ, Mer G, Sixma TK, Richard S. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31:1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette FA, Richard S. JMJD2A promotes cellular transformation by blocking cellular senescence through transcriptional repression of the tumor suppressor CHD5. Cell Rep. 2012;2:1233–1243. doi: 10.1016/j.celrep.2012.09.033. [DOI] [PubMed] [Google Scholar]
- Negoro S, Kunisada K, Fujio Y, Funamoto M, Darville MI, Eizirik DL, Osugi T, Izumi M, Oshima Y, Nakaoka Y, Hirota H, Kishimoto T, Yamauchi-Takihara K. Activation of signal transducer and activator of transcription 3 protects cardiomyocytes from hypoxia/reoxygenation-induced oxidative stress through the upregulation of manganese superoxide dismutase. Circulation. 2001;104:979–981. doi: 10.1161/hc3401.095947. [DOI] [PubMed] [Google Scholar]
- Palomera-Sanchez Z, Bucio-Mendez A, Valadez-Graham V, Reynaud E, Zurita M. Drosophila p53 is required to increase the levels of the dKDM4B demethylase after UV-induced DNA damage to demethylate histone H3 lysine 9. J Biol Chem. 2010;285:31370–31379. doi: 10.1074/jbc.M110.128462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires IM, Bencokova Z, Milani M, Folkes LK, Li JL, Stratford MR, Harris AL, Hammond EM. Effects of acute versus chronic hypoxia on DNA damage response and genomic instability. Cancer Res. 2010;70:925–935. doi: 10.1158/0008-5472.CAN-09-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard PJ, Loenarz C, Mole DR, McDonough MA, Gleadle JM, Schofield CJ, Ratcliffe PJ. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem J. 2008;416:387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Jiménez FJ, Moreno-Manzano V, Lucas-Dominguez R, Sánchez-Puelles JM. Hypoxia causes downregulation of mismatch repair system and genomic instability in stem cells. Stem Cells. 2008;26:2052–2062. doi: 10.1634/stemcells.2007-1016. [DOI] [PubMed] [Google Scholar]
- Shi L, Sun L, Li Q, Liang J, Yu W, Yi X, Yang X, Li Y, Han X, Zhang Y, Xuan C, Yao Z, Shang Y. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci USA. 2011;108:7541–7546. doi: 10.1073/pnas.1017374108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- Slee RB, Steiner CM, Herbert BS, Vance GH, Hickey RJ, Schwarz T, Christan S, Radovich M, Schneider BP, Schindelhauer D, Grimes BR. Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene. 2012;31:3244–3253. doi: 10.1038/onc.2011.502. [DOI] [PubMed] [Google Scholar]
- Sullivan R, Graham CH. Hypoxia prevents etoposide-induced DNA damage in cancer cells through a mechanism involving hypoxia-inducible factor 1. Mol Cancer Ther. 2009;8:1702–1713. doi: 10.1158/1535-7163.MCT-08-1090. [DOI] [PubMed] [Google Scholar]
- Tierney BJ, McCann GA, Cohn DE, Eisenhauer E, Sudhakar M, Kuppusamy P, Hideg K, Selvendiran K. HO-3867, a STAT3 inhibitor induces apoptosis by inactivation of STAT3 activity in BRCA1-mutated ovarian cancer cells. Cancer Biol Ther. 2012;13:766–775. doi: 10.4161/cbt.20559. [DOI] [PubMed] [Google Scholar]
- Toyokawa G, Cho HS, Lwai Y, Yoshimatsu M, Takawa M, Hayami S, Maejima K, Shimizu N, Tanaka H, Tsunoda T, Field HI, Kelly JD, Neal DE, Ponder BA, Maehara Y, Nakamura Y, Hamamoto R. The histone demethylase JMJD2B plays an essential role in human carcinogenesis through positive regulation of cyclin-dependent kinase 6. Cancer Prev Res (Phila) 2011;4:2051–2061. doi: 10.1158/1940-6207.CAPR-11-0290. [DOI] [PubMed] [Google Scholar]
- Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzatsos A, Paskaleva P, Lymperi S, Contino G, Stoykova S, Chen Z, Wong KK, Bardeesy N. Lysine-specific demethylase 2B (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. J Biol Chem. 2011;286:33061–33069. doi: 10.1074/jbc.M111.257667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc Natl Acad Sci USA. 2003;100:12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JA, Watson CJ, McCann A, Baugh J. Epigentics, the epicenter of the hypoxic response. Epigenetics. 2010;5:293–296. doi: 10.4161/epi.5.4.11684. [DOI] [PubMed] [Google Scholar]
- Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, Derecka M, Szczepanek K, Szelag M, Gornicka A, Moh A, Moghaddas S, Chen Q, Bobbili S, Cichy J, Dulak J, Baker DP, Wolfman A, Stuehr D, Hassan MO, Fu XY, Avadhani N, Drake JI, Fawcett P, Lesnefsky EJ, Larner AC. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L, Chen Y, Zeng LL, Zhao F, Li R, Liu Y, Zhang C. Triptolide induces cell-cycle arrest and apoptosis of human multiple myeloma cells in vitro via altering expression of histone demethylase LSD1 and JMJD2B. Acta Pharmacol Sin. 2012;33:109–119. doi: 10.1038/aps.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Zhang ZG, Tian XQ, Sun DF, Liang QC, Zhang YJ, Lu R, Chen YX, Fang JY. Inhibition of JAK1,2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia. 2008;10:287–297. doi: 10.1593/neo.07971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki Y, Kimura H, Hosoi T, Ozawa K. MyD88 plays a key role in LPS-induced Stat3 activation in the hypothalamus. Am J Physiol Regul Integr Comp Physiol. 2010;298:R403–R410. doi: 10.1152/ajpregu.00395.2009. [DOI] [PubMed] [Google Scholar]
- Yang J, Jubb AM, Pike L, Buffa FM, Turley H, Baban D, Leek R, Gatter KC, Ragoussis J, Harris AL. The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res. 2010;70:6456–6466. doi: 10.1158/0008-5472.CAN-10-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LC, McDonald DW, Hendzel MJ. Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following γ-irradiation. J Biol Chem. 2013;288:21376–21388. doi: 10.1074/jbc.M113.491514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Narayanan L, Rockwell S, Glazer PM. Diminished DNA repair and elevated mutagenesis in mammalian cells exposed to hypoxia and low pH. Cancer Res. 2000;60:4372–4376. [PubMed] [Google Scholar]
- Zeng J, Ge Z, Wang L, Li Q, Wang N, Björkholm M, Jia J, Xu D. The histone demethylase RBP2 is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells. Gastroenterology. 2010;138:981–992. doi: 10.1053/j.gastro.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–4139. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chen L, Pledger WJ, Fang J, Chen J.2013p53 promotes repair of heterochromatin DNA by regulating JMJD2b and SUV39H1 expression Oncogenee-pub ahead of print 4 February 2013; doi: 10.1038/onc.2013.6 [DOI] [PMC free article] [PubMed]
- Zhu S, Li Y, Zhao L, Hou P, Shangguan C, Yao R, Zhang W, Zhang Y, Tan J, Huang B, Lu J. TSA-induced JMJD2B downregulation is associated with cyclin B1-dependent survivin degradation and apoptosis in LNCap cells. J Cell Biochem. 2012;113:2375–2382. doi: 10.1002/jcb.24109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.