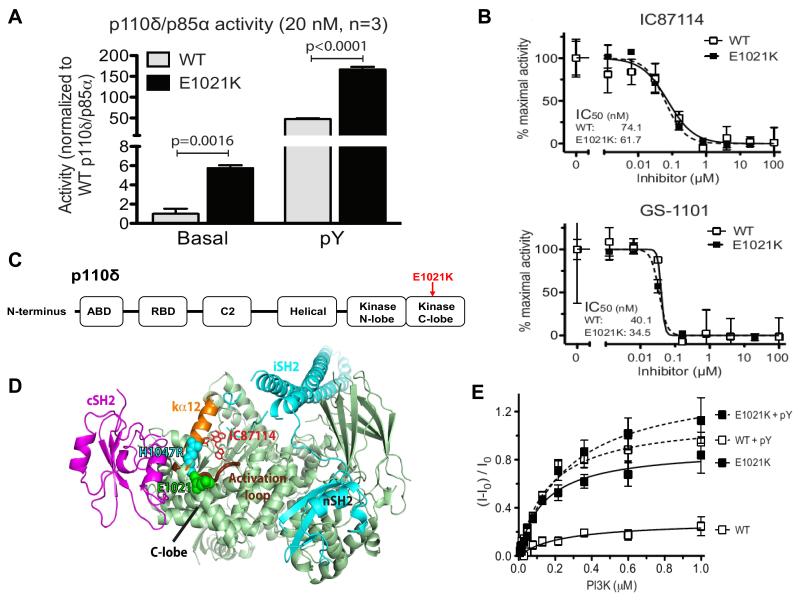
Fig. 2. In vitro activity and structure of p110δ.
(A) Basal and pY-stimulated PI3K activity at 20 nM concentration. Graphs are mean ± SD of 3 independent experiments. P-values were calculated by two-tailed t-test. (B) Inhibition of mutant and wild-type p110δ/p85α as a function of IC87114 or GS-1101 concentration (data are mean ± SD, N=3). (C) Domain organization of p110δ. (D) Structural model of the p110δ/p85α heterodimer. p110δ catalytic subunit (pale green), nSH2 and iSH2 domains of the p85 regulatory subunit (cyan), cSH2 domain (magenta), p110δ activation loop (thick chocolate tube beneath kα12), residue E1021 of p110δ (green spheres) and the analogous residue in H1047R mutant of p110α (cyan spheres). The IC87114 inhibitor bound in the active site is shown in stick representation. (E) Membrane binding of p110δ. FRET between the PI3K complex and Dansyl-PS-containing membrane vesicles in the absence (solid lines) or presence (dashed lines) of the pY peptide (data are mean ± SD, N=3).