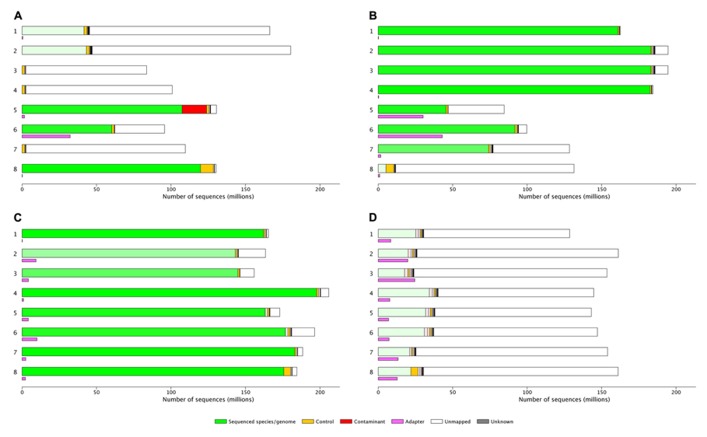
FIGURE 1.
(A) 997M reads, lots of bad libraries and higher than expected PhiX indicating poor quantification of sequencing libraries: Lanes 1 and 2 user A: 25%A, 73%U, 3.5%E, > 1%Ad. Lanes 3,4 and 7 user B: 97%U, these libraries were from a species that was not included in the set of reference genomes aligned to using MGA, 1–2%P. Lanes 5,6 and 8 individual users, lane 5 shows 13% mouse genome contamination, lane 6 35%U, 34%Ad, lane 8 92%A, 0.2%E, 6.6%P. (B) 1180M reads, quality can be attributed to different library preparations. Lanes 1–4 user A: 99%A, low error rate, and no adapter. Lane 5 user B: 53%A, 46%U, 37%Ad (a known problem with this library type). Lane 6 user C: 92%A, 5.8%U, 43%Ad. Lane 7 user D: 59%A, 0.55%E, 38%U. Lane 8 user D 91%U. (C) 1432M read. Lane 1 user A: 98%A, 0.3%E. Lane 2 and 3 user B 87–93%A, 6–11%U, 0.6–0.8%E, 3–6%Ad. Lane 4 user C: 96%A, 0.3%E. Lane 5–8 user D: 90–97%A, 2–8%U, 0.2–0.36%E, 1–5%Ad. (D) 1193M reads, a good flow cell with what look like poor libraries. Lanes 1–8 user A: 11–24%A, 72–84%U, 2.5–3.6%E, 5–16%Ad. %A, percent aligned to the target genome; %U, percent unmapped; %P, percent PhiX control genome; %E, error rate; %Ad, percent aligned to adapter sequences.