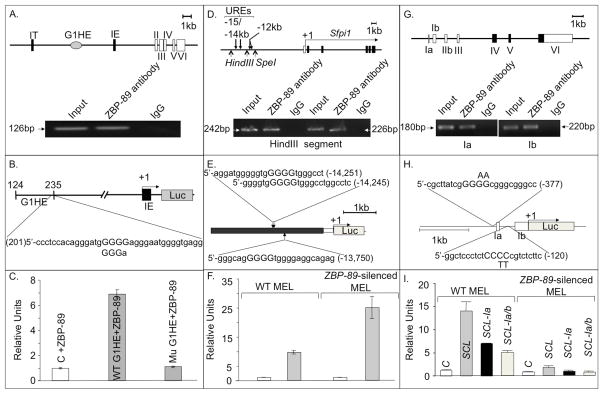
Figure 6. ZBP-89 is a transcriptional regulator of GATA1, PU.1 and SCL.
(A) Schematic of a genomic segment containing mouse GATA1 locus, with two cell-type specific first exons (IT and IE), five coding exons (II–VI), and the G1HE region. The other cis-regulatory elements (double GATA, CACCC and GATA repeats) are not shown. Lower panel, ChIP assays showing specific binding of ZBP-89 to the 5′ region of the GATA1 gene hematopoietic enhancer (G1HE)(which allows GATA1 expression in erythroid lineage). (B) Wt and mutant G1HE (G1HE-(124–235)-luc in which one of the G5 string comprising the ZBP-89 core binding motif is deleted) reporter constructs used (see methods). (C) Histograms (mean±SD, n=2) showing luciferase (Luc) activity in ZBP-89-transfected QM7 cells driven by wild-type (WT) or mutant G1HE (where the ZBP-89 site is mutated). Background luciferase activity was obtained using the control (C) promoter-less vector pGL2. (D) Schematic of a genomic segment containing the nontranslated region of PU.1, the minimal promoter (white bar) and the five coding exons (in black). Transcription start site (+1) and direction (horizontal arrow) are shown. Lower panel, ChIP assays showing specific interaction of ZBP-89 with two PCR products in the −15/−14 URE of PU.1. Normal IgG, used as negative control, and input DNA used as a positive control. (E) pXP-214 kb/-0.334 kb/Luc plasmid containing the 3.5kb −15/−14 URE (in black) cloned upstream of the PU.1 minimal 0.5kb promoter (in white) driving Luc reporter (in gray). Position and sequence of the three predicted ZBP-89 binding sites (core motif capitalized) in −15/14 URE are shown. (F) Histograms (mean±SD, n=2) showing Luc activity in WT and ZBP-89-silenced MEL cells driven by −15/14 URE. In C, F, I, relative units represent Luc activity normalized against β-Gal values. (G) Schematic of a genomic segment containing mouse SCL, comprising 7 exons, with noncoding exons in white. Lower panel, ChIP assays showing specific interaction of ZBP-89 with 1a or 1b promoter regions. (H) Schematic of the construct containing promoters la and lb of SCL cloned upstream of Luc in SCL-pGL2 vector. The two predicted ZBP-89 binding sites are shown. Position and nature of substitution of the two central pyrimidines in the ZBP-89 binding motif of la and lb are indicated above and below the respective sequence. Exons 1a and 1b are shown as white boxes. (I) Histograms (mean ± SD, n=2) showing Luc activity driven by −2kb to +1 DNA region of WT SCL or by SCL in which ZBP-89 consensus-binding sites in promoter 1a (SCL-1a) or 1a+1b (SCL-1a/b) were mutated. Background luciferase activity was obtained using the control promoter-less vector pGL2.