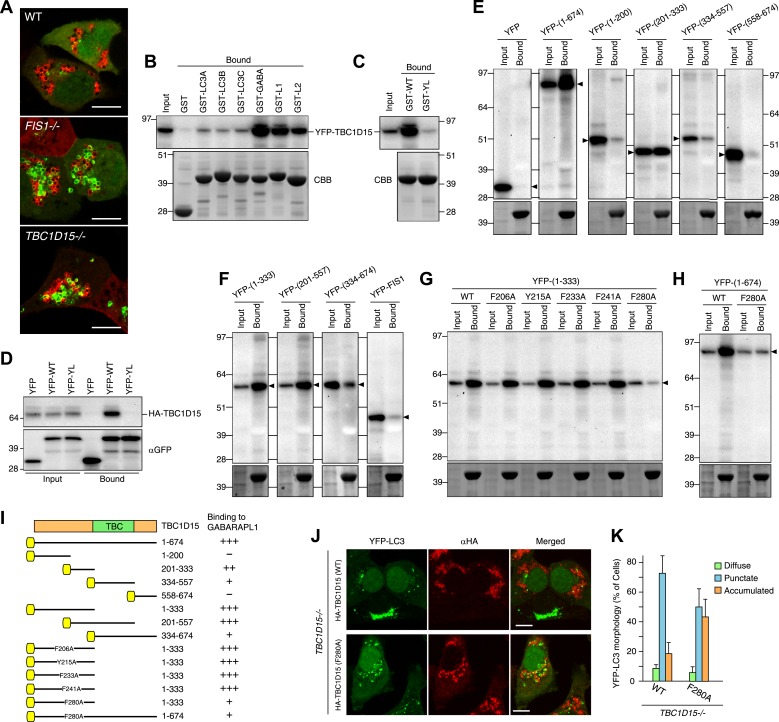
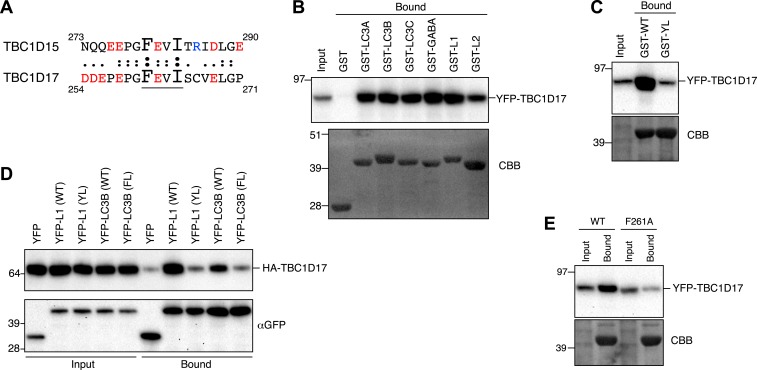
Figure 7. TBC1D15 binds ATG8 family proteins.
(A) The indicated cells transiently expressing YFP-GABARAPL1 (green) and mCherry-Parkin (red) were treated with valinomycin for 3 hr. Scale bars, 10 μm. (B) YFP-TBC1D15 overexpressed in HEK293 cells was subjected to binding assays with GST-fused proteins (GABA, L1, and L2 represent GABARAP, GABARAPL1, and GABARAPL2, respectively). 5% input and bound fractions were analyzed by immunoblotting with anti-GFP antibody (upper panel). Coomassie brilliant blue (CBB) staining shows GST-fusion proteins in bound fractions (lower panel). (C) Binding assay carried out as in (B) with GST-GABARAPL1 WT (GST-WT) or its Y49A/L50A mutant (GST-YL). Immunoblotting with anti-GFP antibody (upper panel) and CBB staining (lower panel) are shown. (D) Cell extracts from HEK293 overexpressed HA-TBC1D15 and YFP, YFP-GABARAPL1 (YFP-WT), or its Y49AL50A mutant (YFP-YL) were subjected to pull down assays with GFP-Trap. 5% input and bound fractions were analyzed by immunoblotting with anti-HA (upper panel) and anti-GFP (lower panel) antibodies. (E–H) The indicated YFP-tagged TBC1D15 full-length, truncated, or point-mutant protein or YFP-Fis1 overexpressed in HEK293 cells were subjected to binding assays with recombinant GST-GABARAPL1. 5% input and bound fractions were analyzed by immunoblotting with anti-GFP antibody (upper panel) and CBB staining (lower panel). (I) Summary of binding abilities of truncated or point-mutated TBC1D15 constructs. –, +, ++, and +++ indicates binding of recombinant GST-GABARAPL1 to less than 1%, 1–5%, 5–10%, and over 10%, respectively of the total YFP-TBC1D15 fragment. Yellow boxes indicate YFP tags. (J) YFP-LC3 and mCherry-Parkin stably expressing TBC1D15−/− cells in the presence of HA-tagged TBC1D15 WT or F280A mutant were treated with valinomycin for 3 hr. Cells were subjected to immunofluorescence microscopy with anti-HA antibody. Scale bars, 10 μm. (K) The YFP-LC3 morphology of cells in (J) was quantified. The error bars represent ±SD from three independent replicates. Over 50 cells were counted in each well.