Abstract
Like all cancers, brain tumors require a continuous source of energy and molecular resources for new cell production. In normal brain, glucose is an essential neuronal fuel, but the blood-brain barrier limits its delivery. We now report that nutrient restriction contributes to tumor progression by enriching for brain tumor initiating cells (BTICs) due to preferential BTIC survival and adaptation of non-BTICs through acquisition of BTIC features. BTICs outcompete for glucose uptake by co-opting the high affinity neuronal glucose transporter, type 3 (Glut3, SLC2A3). BTICs preferentially express Glut3 and targeting Glut3 inhibits BTIC growth and tumorigenic potential. Glut3, but not Glut1, correlates with poor survival in brain tumors and other cancers; thus, TICs may extract nutrients with high affinity. As altered metabolism represents a cancer hallmark, metabolic reprogramming may instruct the tumor hierarchy and portend poor prognosis.
Introduction
Glioblastomas (GBM; World Health Organization grade IV gliomas) are the most lethal and prevalent primary malignant brain tumors in adults with median survival remaining 14.6 months with the best available therapies1. The inability to effectively manage GBMs has motivated the search for more effective treatments. Recent observations underscore the importance of inter- and intratumoral heterogeneity driven by genetic and non-genetic causes to therapeutic responses and patient outcomes. Heterogeneity within the neoplastic compartment is partially explained by the tumor initiating cell (TIC) hypothesis that holds that a cellular hierarchy exists in some cancers with self-renewing TICs generating progeny constituting the tumor bulk2. Although the TIC hypothesis remains controversial, multiple groups, including our own, have demonstrated that brain tumor initiating cells (BTICs) express stem cell markers, display sustained self-renewal, differentiate towards multiple lineages, and phenocopy the original tumor upon xenotransplantion3-6. BTICs also display radio- and chemoresistance, which is thought to contribute to tumor recurrence following treatment5,6. Thus, targeting of BTICs offers a potential paradigm for GBM control.
Nutrient acquisition and utilization are critical for growth of tumors, and metabolic alterations in cancers are recognized as the Warburg Effect7: the observation that cancer cells become less reliant on oxygen-dependent mitochondrial oxidative phosphorylation and instead rely on the anaerobic but glucose-intensive glycolysis pathway for ATP generation. This metabolic reprogramming generates glycolytic end products necessary to produce biological building blocks (proteins, nucleic acids, and lipids) required for tumor growth even under hypoxia. As ATP production per glucose molecule is lower with anaerobic metabolism, tumors ultimately require higher glucose flux than normal tissues. The brain is an extremely metabolically active organ that derives energy almost entirely from glucose, and the lack of extensive energy stores in the brain necessitates tight control of blood glucose homeostasis8. However, the difference in glucose uptake in normal and neoplastic brain is complex and has been exploited clinically with [18F]-deoxyglucose PET (positron emission tomography) imaging. The clinical importance of glucose consumption for brain tumor growth is also suggested by reports indicating higher glucose levels in brain tumor patients associate with shorter survival9. Vascular glucose delivery to the normal brain is physiologically stymied by the blood-brain barrier. In response, neurons express the specialized glucose transporter isoform, type 3 (Glut3). Glut3 has a five-fold higher affinity for glucose than the ubiquitous glucose transporter, type I (Glut1), and Glut3 expression is largely restricted to cells with both a high glucose demand and a glucose-poor microenvironment. Cancer glucose uptake is thought to be primarily driven by Glut1. Limited reports demonstrate Glut3 expression in cancers, but its functional importance has been largely ignored. Recently, the concepts of metabolic reprogramming and oncogenic metabolites support key roles of metabolism during transformation with a similar transition to a glycolytic state during somatic cell reprogramming10,11. These data and others suggest the importance of understanding the mechanisms driving metabolic adaptation in cancer and specifically within the BTIC fraction to develop novel treatments.
In GBMs and other solid tumors, glucose metabolism is elevated in microenvironmental conditions associated with poor vascular supply such as hypoxia and reduced extracellular pH. BTICs are enriched in areas of necrosis12. Both hypoxia13 and acidic stress14 induce increased BTIC functional readouts: neurosphere formation and tumorigenic potential. These data suggest that regulation of metabolic processes and resulting changes in the tumor microenvironment have significant effects on the BTIC phenotype. As reduced blood flow in growing tumors can cause localized nutrient deprivation with very low levels of glucose, we considered whether molecular differences in BTICs permit improved competition for limited resources.
Results
Nutrient Restriction Promotes a BTIC Phenotype
To determine if glucose deprivation influences TICs, we exposed bulk GBM cells to media containing standard (4.5 g/L) or restricted (0.45 g/L) levels of glucose. The clinical equivalent of standard media glucose levels is 450 mg/dL, high above normal (70-144 mg/dL). Blood glucose levels higher than 200 mg/dL are considered hyperglycemic, but not extraordinary in GBM patients whose mean glucose levels can range as high as 459 mg/dL9. Brain glucose levels in animal models are 15-20% of blood levels15-16, suggesting that brain glucose can range from 15-90 mg/dL. The lower range of GBM patient mean serum glucose levels is 65 mg/dL9, but glucose levels in interstitial fluid from solid tumors are below 25 mg/dL16. Together, these data indicate that the “restricted” levels of glucose used for the studies presented here are representative of physiological conditions, whereas “normal” cell culture conditions represent severe hyperglycemia.
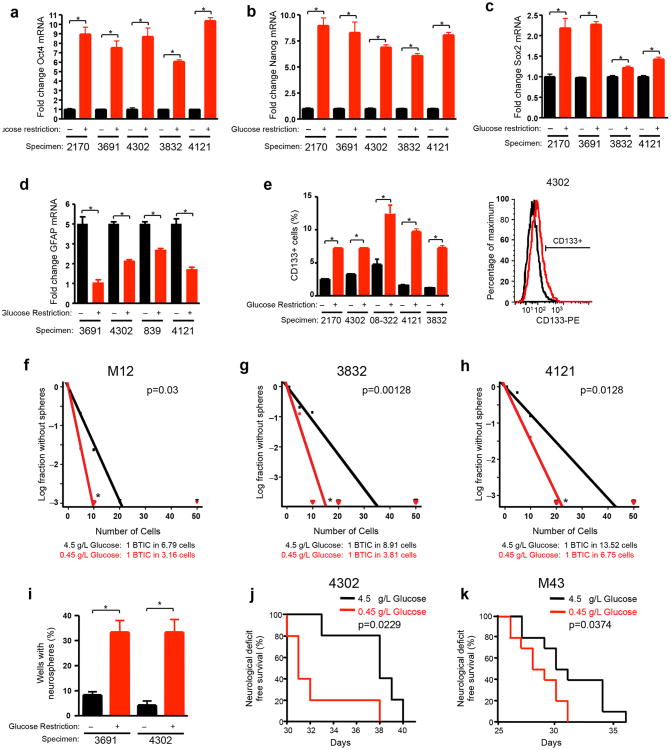
As cultures prospectively enriched for BTICs express stem cell markers, we first analyzed the expression of a subset of BTIC markers in non-enriched (bulk) GBM cells derived from human patient tumors cultured under standard or restricted glucose for one week. Quantitative real-time PCR of mRNA demonstrated six- to ten-fold elevation of the core stem cell transcription factors markers Oct4 (Fig. 1a) and Nanog (Fig. 1b) in the cultures with restricted glucose. Sox2 expression was also consistently, but more modestly, increased with a greater than two-fold increase in half the samples tested (Fig. 1c). Levels of the astrocyte differentiation marker glial fibrillary protein (GFAP) decreased with low glucose culture (Fig. 1d). Using flow cytometry, we observed a two-fold or greater increase in the percentage of cells expressing a putative BTIC immunophenotype [the cell surface marker CD133 (Prominin1)] after exposure to low glucose (Fig. 1e). These data demonstrate that glucose deprivation increases the expression of multiple BTIC markers and decreased differentiation.
Figure 1. Glucose Restriction Promotes a Brain Tumor Initiating Cell Phenotype.
Unenriched glioma cells isolated from the indicated xenografts were cultured for 7 days in standard (4.5 g/L) or restricted glucose (0.45 g/L). Quantitative real-time PCR indicates elevation (a) Oct4 [*, p<0.0001 with unpaired t-test], (b) Nanog [*, p<0.0001 with unpaired t-test], and (c) Sox2 [*, p<0.05 with unpaired t-test] mRNA under conditions of restricted glucose in comparison to standard culture conditions while (d) GFAP is reduced [*, p<0.05 with unpaired t-test]. (e) Flow cytometry demonstrates increased percentages of CD133+ cells after culture of unenriched tumor cells in low glucose [*, p<0.008 with unpaired t-test]. An overlay of representative flow plots is shown at right. (f-h) In vitro limiting dilution assays plating decreasing numbers of bulk tumor cells from (f) M12 [p=0.03 with ELDA analysis], (g) 3832 [p=0.00128 with ELDA analysis] or (h) 4121 [p=0.0128 with ELDA analysis] GBM cells indicate the frequency of BTICs increases after culture in low glucose. (i) Neurosphere formation assays indicate the percentage of wells with neurospheres increases after culture of unenriched GBM cells in restricted glucose conditions when 10 cells are plated per well [*, p<0.007 with unpaired t-test]. Kaplan Meier survival curves of immunocompromised mice intracranially injected with (j) 4302 [n=5 per arm; *p<0.05 with log-rank analysis] or (k) M43 [n=10 per arm; *p<0.05 with log-rank analysis] cells cultured in restricted glucose or standard glucose conditions. *p<0.05 with log-rank analysis.
As TICs are defined functionally, we next determined the effects of restricted glucose in BTIC functional assays. Prospectively enriched BTIC cultures have an increased capacity to form tumorspheres, a phenotype associated with self-renewal and clinical outcomes in GBM17. In vitro limiting dilution assays using cells isolated from GBM xenografts demonstrated that the frequency of BTICs capable of forming neurospheres increased after culture under glucose restriction (Figs. 1f-h). With a constant number of glucose deprived cells, the percentage of wells with neurospheres also increased three-fold (Fig. 1i). The ability to propagate tumors in vivo is the gold standard for BTIC function. We determined whether culture in low glucose influenced tumorigenic potential using an orthotopic xenograft model. Low glucose culture significantly increased in vivo tumor growth relative to standard glucose culture as demonstrated by reduced median survival and/or increased tumor incidence (Figs. 1j-k; Supplemental Table 1). Xenografts phenotypically recapitulated the histology of the original tumors, demonstrating the BTIC-associated property of tumor propagation (data not shown). These data demonstrate that glucose deprivation promotes the BTIC phenotype .
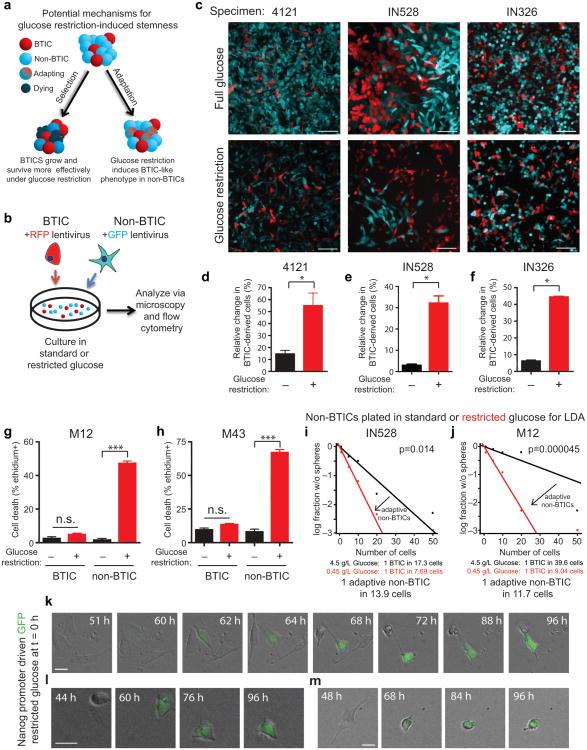
To determine the biological mechanism through which nutrient deprivation enriches for BTICs, we investigated two potential hypotheses: 1) BTICs preferentially survive low glucose conditions, and/or 2) non-BTICs adapt to low glucose by acquiring a more BTIC-like phenotype (Fig. 2a). We generated fluorescently-labeled GBM cells and isolated differentially labeled BTICs and non-BTICs by flow cytometry for use in co-culture experiments (Fig. 2b). We co-cultured cells under standard or low glucose conditions at defined and biologically relevant percentages (i.e. 10% BTIC:90% non-BTIC for 4121 and IN326 or 50% BTIC:50% non-BTIC for IN528) and monitored the percentage of cells and their progeny over time. Imaging demonstrated strong differences in the percentage of BTIC-derived cells under restricted glucose conditions (Fig. 2c). Using flow cytometry, we confirmed that there was a significantly increased percentage change in the fraction of BTICs and their derivatives under glucose restriction (Fig. 2d-f).
Figure 2. Enrichment for Brain Tumor Initiating Cells (BTICs) Under Nutrient Deprivation is Associated with Preferential BTIC survival and Adaptation of non-BTICs Towards a BTIC Phenotype.
(a) Schematic of potential mechanisms for glucose restriction induced BTIC enrichment shows the potential for low glucose to select for surviving BTICs or for non-BTICs to adapt to nutrient restriction through acquisition of BTIC-like phenotypes. (b) Strategy for competition experiments with fluorescently labeled BTICs and non-BTICs. (c) Fluorescently labeled BTICs and non-BTICs were plated at defined numbers via flow cytometry and the percentage of cells derived from them monitored over time in standard (4.5 g/L) or restricted glucose (0.45 g/L). Scale bars represent 75 μm. Representative images of cells after 7 days are shown. Flow cytometry was used to quantify the percentage of BTIC and non-BTIC cells from (d) 4121 [*, p=0.0211 with unpaired t-test], (e) IN528 [*, p<0.02 with unpaired t-test], or (f) IN326 [*, p<0.0005 with unpaired t-test] specimens in standard and low glucose and the relative change in the percentage of BTIC-derived cells is shown. Ethidium homodimer III staining demonstrated elevated cell death in non-BTICs derived from (g) M12 [*, p=0.0011 with unpaired t-test] or (h) M43 [*, p<0.001 with unpaired t-test] GBMs after nutrient restriction. In vitro limiting dilution assays in which non-BTICs from (i) IN528 [p=0.014 with ELDA analysis] or (j) M12 [p=0.000045 with ELDA analysis] were plated directly into normal or low glucose demonstrated adaptation to nutrient restriction through promotion of a BTIC phenotype. (k-m) Time lapse imaging using reporter cells in which the Nanog promoter is driving GFP demonstrated that Nanog/GFP- cells can begin expressing Nanog/GFP after 60 hours of nutrient restriction. Three different panels of time lapse images for individual cells which became GFP+ are shown. Scale bars represent 25 μm.
To further define the mechanism responsible for the changes observed in co-culture experiments, we investigated the survival of BTICs and non-BTICs in normal and low glucose. When ethidium homodimer III staining was used to determine the percentage of dead cells in matched GBM fractions, we did not observe significant increases in BTIC cell death under restricted glucose (Fig. 2g, 2h). However, the fraction of dead BTICs was consistently slightly elevated (Fig. 2g, 2h). In striking contrast, non-BTICs displayed a greater than six-fold increase in dead cells (Fig. 2g, 2h). Together, these data demonstrate that BTICs preferentially survive restricted glucose conditions.
BTIC markers, including Nanog, were elevated in non-BTICs that survived low glucose similar to our observation with bulk tumor cells (data not shown). These data suggested that non-BTICs surviving low glucose adapt through acquisition of a more BTIC-like state. We therefore performed modified in vitro limiting dilution assays with non-BTICs plated directly into wells containing normal or low glucose stem cell media (Fig. 2i, 2j). After an initial incubation period of seven days, we restored normal glucose levels and neurosphere formation subsequently determined after two additional weeks. Using this strategy, we excluded the potential for BTIC enrichment due to selection to promote neurosphere formation capacity. We continued to observe a significant increase in the frequency of sphere forming GBM cells with low glucose exposure (Fig. 2i, 2j).
In an alternative approach, we evaluated the adaptation of non-BTICs to low glucose at the single cell level using time lapse microscopy with reporter cells expressing NANOG-promoter driven GFP. Imaging demonstrated the presence of infrequent NANOG-promoter/GFP negative GBM cells that acquired NANOG expression after more than 60 hours of culture in low glucose (Fig. 2k-m; Supplemental Video 1). Over the same time course, we did not observe NANOG-promoter/GFP expression under normal glucose (data not shown). These data demonstrate that non-BTICs can adapt to nutrient restriction through BTIC marker expression.
Preferential Glucose Uptake by BTICs
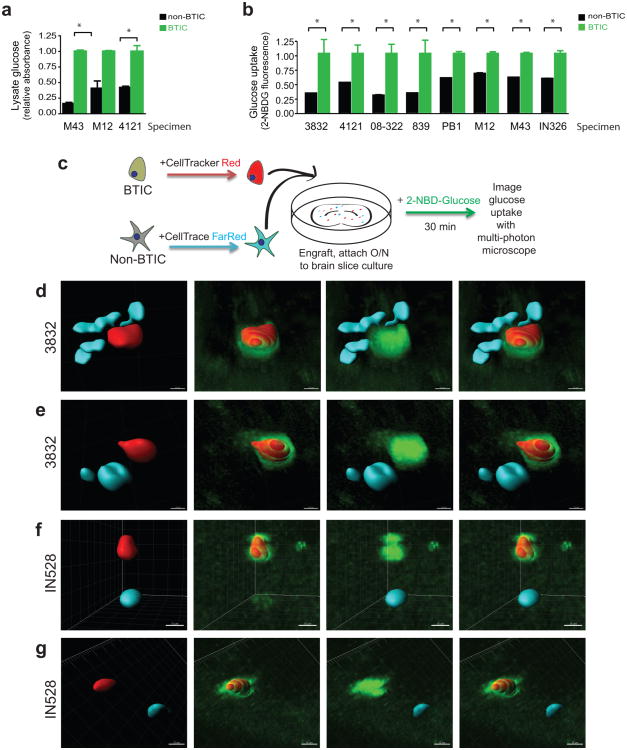
De novo synthesis of molecular building blocks for cell growth requires glucose for energy production18. Highly efficient glucose uptake could therefore provide a competitive advantage contributing to tumor maintenance, particularly with restricted glucose levels. A glucose oxidase colorimetric assay after a pulse post nutrient starvation to measure glucose in cell lysates demonstrated BTICs had higher levels of glucose than matched non-BTICs (Fig. 3a). To directly compare the uptake of glucose in GBM tumor cell subsets, we used a fluorescent glucose analog, 2-[N-(7-nitrobenz-2-oxa-1,3-diaxol-4-yl)amino]-2-deoxyglucose (2-NBDG) that mimics glucose19. We incubated bulk GBM cells with 2-NBDG and an antibody against the BTIC marker CD133. Flow cytometry demonstrated that CD133 expressing cells consistently displayed higher 2-NBDG fluorescence (Fig. 3b). Ex vivo multiphoton imaging of labeled GBM subpopulations in brain slice cultures confirmed that BTICs outcompete for glucose with surrounding brain and differentiated tumor cells (Fig. 3c-g, Supplementary Fig. 1a). Quantification of fluorescence visualized through multiphoton imaging with Imaris reconstruction20 demonstrated that the uptake of NBDG was greater than two-fold higher in the BTICs (Supplementary Figs. 1b-c), consistent with results in the flow cytometry assays. Together, these data indicate that BTICs preferentially uptake glucose relative to their non-BTIC counterparts.
Figure 3. Brain Tumor Initiating Cells Preferentially Uptake Glucose.
(a) Glucose oxidase assays demonstrate elevated levels of glucose in the lysate of BTICs in comparison to matched non-BTICs [*, p<0.05 with unpaired t-test]. (b) Flow cytometry demonstrates uptake of a fluorescent glucose analogue is higher in CD133 expressing glioma cells [*, p<0.001 with unpaired t-test]. (c) Diagram of the experimental procedure for ex-vivo imaging of glucose uptake in brain slices. CD133-sorted BTICs or non-BTICs were labeled for 45 minutes with CellTracker Red CMPTX or CellTrace FarRed DDAO-SE, respectively. Cells were then engrafted onto live mouse brain slice cultures and allowed to engraft overnight. The next day, the slice culture with engrafted cells was incubated for 30 minutes in 50ug/ml 2-NBDG and imaged via multiphoton microscopy. (d-g) Reconstructions of representative fields, showing increased glucose (green) uptake in BTICs (red). Non-BTICs (cyan) consistently displayed less glucose uptake than BTICs. Scale bars represent 10 μm for d and 15 μm for e-g for the window of the 3D reconstruction.
BTICs Express Elevated Glucose Transporter Type 3
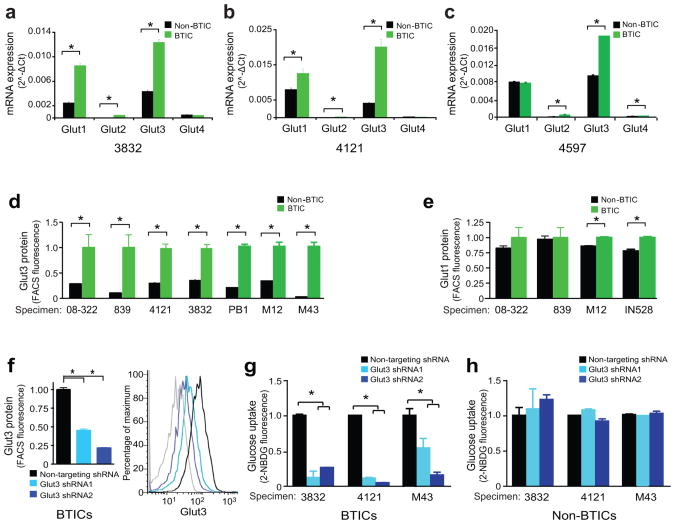
Cellular uptake of glucose occurs through facilitated diffusion using a family of solute carriers, the Glut family of proteins. The major Glut isoforms in normal brain are Glut1 and Glut3, which are differentially expressed: Glut1 is broadly expressed in both glia and neurons, whereas the higher affinity Glut3 is restricted to neurons21 (Supplementary Fig. 2a, 2b). Limited prior reports on Glut expression in brain tumors including GBMs also indicate that Glut1 and Glut3 are likely to be the predominant isoforms expressed22,23. To determine whether differential Glut isoform expression in BTICs contributes to preferential glucose uptake, we examined the mRNA levels of glucose transporter isoforms. As Glut1 and Glut3 expression increase with extended cell culture (Supplementary Fig. 2c, 2d), we performed experiments within five passages following tumor dissociation. Glut1 and Glut3 expression was observed in both BTICs and non-BTICs with minimal Glut2 and Glut4 levels (Figs. 4a-c). Glucose transporter isoform expression was most significantly different for Glut3 (Figs. 4a-c), but we often observed significantly increased mRNA levels of Glut1 as reported (Figs. 4a-b)12. To extend these observations of mRNA difference into protein analysis, we examined Glut3 (Fig. 4d) and Glut1 (Fig. 4e) expression via flow cytometry. Fluorescent intensity of Glut3 (Fig. 4d) was consistently elevated in BTICs in comparison to matched non-BTICs. Protein expression of Glut1 (Fig. 4d) was sometimes increased in BTICs to a level of statistical significance, but the biological significance of this difference in Glut1 expression is likely to be less than for Glut3 (Fig. 4e). We observed a 20% change in Glut1 expression between BTICs and non-BTICs, whereas there was 300% change in Glut3 expression (Fig. 4d-e). We also evaluated the expression of Glut3 and the BTIC marker Sox2 using co-immunofluorescence (Supplementary Fig. 2e). In bulk GBM cultures, we observed co-localization of Glut3 and Sox2 suggesting Glut3 protein is expressed in BTICs (Supplementary Fig. 2e). These data suggest that Gluts can be differentially expressed in the brain tumor hierarchy, potentially contributing to differences in glucose uptake efficiency.
Figure 4. Increased Expression of Glut3 in Brain Tumor Initiating Cells Drives Glucose Uptake.
mRNA Expression of glucose transporters Glut1, Glut2, Glut3, and Glut4 in BTICs and non-BTICs isolated from (a) 3832 [*, p<0.02 with unpaired t-test], (b) 4121 [*, p<0.001 with unpaired t-test], or (c) 4597 [*, p<0.0001 with unpaired t-test] GBM cells demonstrates elevated Glut3 in BTICs. Flow cytometry using fluorescently labeled antibodies against (d) Glut3 [*, p<0.0001 with unpaired t-test] or (e) Glut1 [*, p<0.0001 with unpaired t-test] demonstrates elevated Glut3 protein in BTICs. (f) Flow cytometry confirms shRNAs directed against Glut3 (shRNA1 and shRNA2) can reduce the expression of Glut3 protein relative to a non-targeting control shRNA [*, p<0.001 with unpaired t-test]. In the representative flow histogram, the grey line is representative of the isotype control. (g) Knockdown of Glut3 shRNA in BTICs isolated from 3832, 4121, or M43 reduces the uptake of a fluorescent glucose analogue demonstrating a requirement for Glut3 in BTIC glucose transport [*, p<0.005 with ANOVA comparison]. (h) There is no requirement for Glut3 for glucose uptake in matched non-BTICs. To best determine the impact of Glut3 directed shRNAs in g-h, values were normalized to non-targeting shRNAs in each group.
Glucose Transporter Type 3 Mediates Glucose Uptake in BTICs
To functionally demonstrate the importance of Glut3 in BTIC glucose uptake, we utilized a targeting approach based on lentivirally expressed shRNAs to reduce levels of Glut3 mRNA (Fig. 4f and data not shown). Targeting of Glut3 had no effect on Glut1 expression (Supplementary Fig 2f). Expression of Glut3 shRNA in BTICs significantly impaired uptake of 2-NBDG in comparison to the non-targeting control (Fig. 4g). In contrast, there was no significant effect of Glut3 knockdown on glucose uptake in matched non-BTICs (Fig. 4h). The data demonstrate that Glut3 is preferentially required for the elevated uptake of glucose in BTICs.
Glut3 shRNA Reduces BTIC Growth and Tumorigenic Potential
We next evaluated the phenotypic consequences of reduced uptake of glucose in BTICs with impaired Glut3 expression. Knockdown of Glut3 significantly decreased the growth of BTICs (Fig. 5a, 5b), while there was no effect on non-BTICs (Supplemental Fig. 2g, 2h), further demonstrating the preferential requirement for Glut3 in BTICs. To determine if targeting of Glut3 also influenced neurosphere formation potential, we quantified BTIC frequency by in vitro limiting dilution in cells expressing non-targeting or Glut3 directed shRNAs. Knockdown of Glut3 resulted in a six-fold or greater decrease in the frequency of BTICs (Figs. 5c, 5d). To further determine whether Glut3 targeting resulted in reduced tumor propagation in vivo, we implanted BTICs expressing non-targeting control shRNA or two different Glut3 directed shRNAs into the right frontal lobes of immunocompromised mice. Daily monitoring of recipient mice for the development of neurological signs (e.g. lethargy, ataxia, seizures and/or paresis) as an indicator of GBM growth demonstrated significantly improved deficit-free survival with Glut3 knockdown in BTICs (Figs. 5e-i). In addition, tumor incidence was reduced with Glut3 shRNA in comparison to the non-targeting control (Figs. 5e-i; Supplemental Tables 2-4). Thus, Glut3 function maintains BTICs.
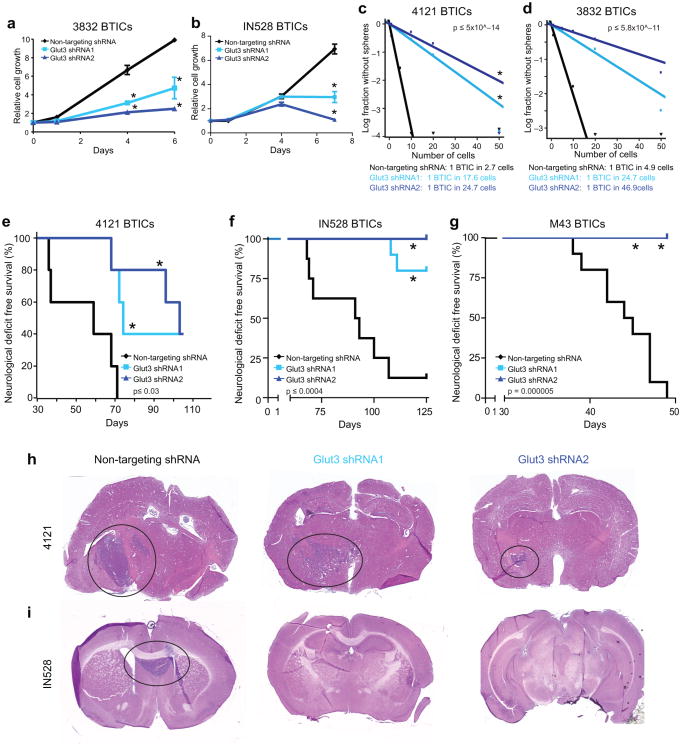
Figure 5. Glut3 Regulates the Brain Tumor Initiating Cell Growth and Self-Renewal.
shRNA directed against Glut3 (shRNA1 and shRNA2) decreases the growth of (a) 3832 [*, p<0.05 with ANOVA] and (b) IN528 [*, p<0.001 with ANOVA] BTICs in comparison to a non-targeting control shRNA as measured using the cell-titer assay. In vitro limiting dilution assays demonstrate that knockdown of Glut3 in BTIC enriched cultures decreases the frequency of (c) 4121 [p=5×10−14 with ELDA analysis] or (d) 3832 [p=5.8×10−11 with ELDA analysis] BTICs. (e) In vivo tumor propagation assays with classical subtype 4121 derived BTICs demonstrate that targeting of Glut3 using shRNA increases the survival of mice bearing human glioma xenografts relative to the non-targeting shRNA control. [n=5 for all groups; * p<0.03 with log-rank analysis]. (f) In vivo tumor propagation assays with proneural subtype IN528 derived BTICs demonstrate that targeting of Glut3 using shRNA increases the survival of mice bearing human glioma xenografts relative to the non-targeting shRNA control [n=8 for non-targeting control, n=10 for Glut3 shRNA1 and Glut3 shRNA2 groups; * p<0.0004 with log-rank analysis]. (g) In vivo tumor propagation assays with classical subtype M43 derived BTICs demonstrate that targeting of Glut3 using shRNA increases the survival of mice bearing human glioma xenografts relative to the non-targeting shRNA control [n=10 for all groups; * p=0.000005 with log-rank analysis]. (h, i) Representative images of hematoxylin and eosin stained sections of brains from mice in e and f demonstrating the presence of tumors in animals showing neurologic signs.
Glut3 Correlates with Brain Tumor Clinical Outcomes
To confirm the clinical relevance of Glut3 expression, we interrogated available brain tumor datasets for Glut isoforms, including Glut3 (Fig. 6a; Supplementary Figs. 3-5). We identified several annotated brain tumor data sets in which we could compare Glut isoform expression in specimens with different tumor grade or recurrence or from patients with differential survival outcomes24-27. Glut3 was strongly correlated with tumor grade in two datasets (Fig. 6b, Fig. 6c, Supplementary Figs. 3a, 4a, 5a). Neither Glut2 nor Glut4 correlated with grade while Glut1 slightly correlated with tumor grade in one dataset (1.1 fold, Fig. 6c), while the fold change in Glut3 expression was higher (1.68 fold) and strongly significant. Glut3 levels are increased upon disease recurrence (Fig. 6d), unlike other Gluts (Supplementary Figs. 3b, 4b, 5b). Further, only Glut3 was a negative prognostic factor for long term (>3 year) survival (Fig. 6e, Supplementary Figs. 3c, 4c, 5c). Glut3 expression three-fold higher in tumors isolated from patients dead within three years compared to those who survived, whereas there was no significant difference in the expression of Glut1, Glut2 or Glut4 isoforms (Fig. 6e, Supplementary Figs. 3c, 4c, 5c). Glut3 was also the only glucose transporter isoform which correlated with poor survival at one year (data not shown).
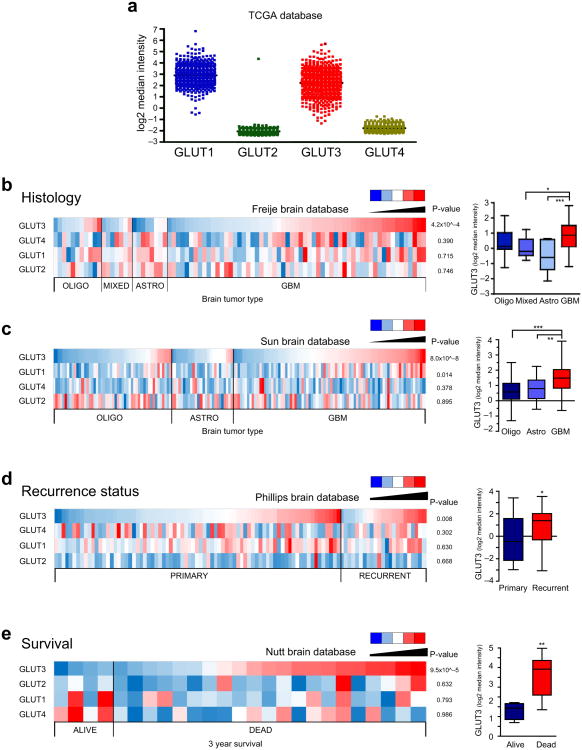
Figure 6. Glut3 Expression is Significantly Increased in Glioblastomas, Recurrent Brain Tumors, and Brain Tumors from Patients with Poor Survival.
(a) Expression of Glut isoforms in the TCGA database demonstrates expression of both Glut1 and Glut3 with minimal expression of Glut2 and Glut4. (b) Oncomine analysis of the Freije database indicates elevated Glut3 mRNA expression (but not Glut1, 2, or 4) correlates with increased glioma tumor grade [n=11 oliogodendroglioma; n=7 mixed glioma; n=8 astrocytomas; n=59 GBM; p=4.2×10ˆ−4 with ANOVA]. (c) Oncomine analysis of the Sun database indicates elevated Glut3 mRNA expression correlates with increased glioma tumor grade [n=50 oligodendroglioma; n=26 astrocytoma; n=81 GBM; p=8.02×10ˆ−8 with ANOVA]. There is also a significant correlation with Glut1 mRNA expression [p=0.014] but not Glut2 or Glut4. (d) Oncomine analysis of the Phillips databases indicates elevated Glut3 mRNA expression (but not Glut1, 2, or 4) correlates with glioma recurrence [n=77 primary; n=23 recurrent; p=0.008 with unpaired t-test]. (e) Oncomine analysis of the Nutt database indicates elevated Glut3 mRNA expression (but not Glut1, 2, or 4) correlates with reduced patient survival at three years [n=4 alive; n=21 dead; p=9.5×10ˆ−5 with unpaired t-test].
To further evaluate the potential correlation of Glut3 expression with patient outcome, we generated Kaplan-Meier survival curves from from the Freije, Phillips, and Nutt datasets24,25,27 as well as the the NCI REpository for Molecular BRAin Neoplastic Data (REMBRANDT)28 and The Cancer Genome Atlas (TCGA) 29. In all datasets, Glut3 expression informed poor prognosis (Figs. 7a-e) whereas Glut1, Glut2, or Glut4 did not correlate with patient outcomes (Supplementary Figs. 3d-h, 4d-h, 5d-h). To determine if Glut3 was more closely associated with prognosis in one of the GBM subtypes, data in the TCGA was segregated based on Verhaak classifications30. While we did not observe a significant association of Glut3 with survival in Mesenchymal tumors (Fig. 7f), Glut3 correlated with survival in Classical (Fig. 7g) and Proneural (Fig. 7h) subtypes. Glut1, Glut2, or Glut4 did not correlate with survival in any GBM subtype (Supplementary Figs. 3i-k, 4i-k, 5i-k). Together these data demonstrate that among the Glut isoforms, Glut3 expression is the only consistent indicator of brain tumor patient outcomes.
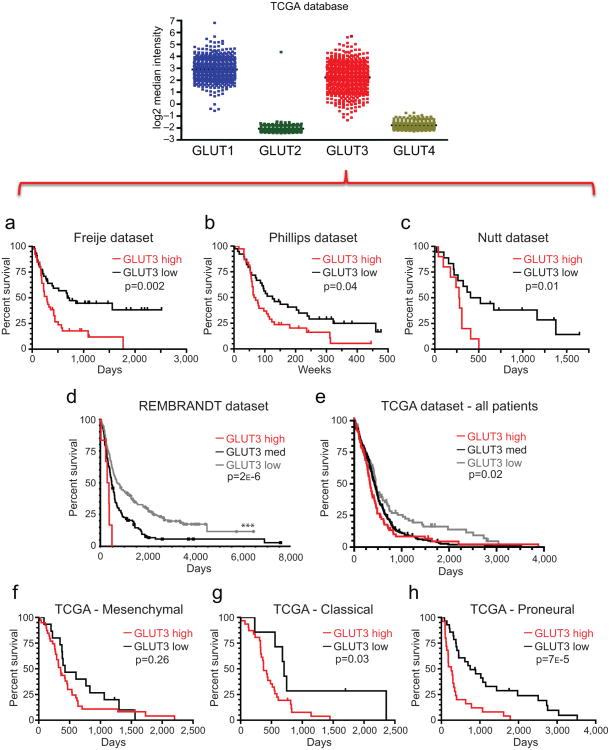
Figure 7. Glut3 Expression Correlates with Poor Survival in Multiple Brain Tumor Datasets.
Analysis of Brain Datasets available through Oncomine indicates a significant correlation between high Glut3 expression and poor survival in the (a) Freije [n=39 Glut3 low; n=45 Glut3 high; p=0.002 with log-rank analysis], (b) Phillips [n=39 Glut3 low; n=38 Glut3 high; p=0.04 with log-rank analysis], and (c) Nutt [n=18 Glut3 low; n=10 Glut3 high; p=0.01 with log-rank analysis] datasets. (d) Analysis of REMBRANDT data in the National Cancer Institute's repository indicates that greater than two fold elevation of Glut3 mRNA expression correlates with poor glioma patient survival [n=193 Glut3 low; n=144 Glut3 medium; n=6 Glut3 high; p=0.002 vs. all other samples and p=0.018 vs. intermediate expression with log-rank analysis]. Greater than two fold reduction in Glut3 mRNA expression correlates with improved patient survival [**, p=2.2×10−6 vs. all other samples and p=7.7×10−6 vs. intermediate expression with log rank analysis]. Reporter is Affymetrix 202499_s_at (Highest Geometric Mean Intensity). (e) Analysis of TCGA data indicates a significant correlation between reduced Glut3 expression and survival in all patients [n=103 Glut3 low; n=320 Glut3 medium; n=91 Glut3 high; *, p=0.02 with log-rank analysis]. Classification of TCGA according to Verhaak subtypes indicates Glut3 does not significantly correlate with survival in (f) Mesenchymal tumors [n=15 Glut3 low; n=39 Glut3 high; p=0.26 with log-rank analysis], but reduced Glut3 correlates with improved patient survival in both (g) Classical [n=7 Glut3 low; n=31 Glut3 high; p=0.03 with log-rank analysis] and (h) Proneural [n=28 Glut3 low; n=25 Glut3 high; p=7×10−5 with log rank analysis] GBM subtypes.
The association between Glut3 and survival was the strongest in the Proneural GBM subtype (Fig. 7h) and this tumor subgroup is highly associated the with glioma-CpG island methylator phenotype (GCIMP)31 and mutational status of the metabolic enzyme isocitrate dehydrogenase (IDH1)32. Glut3 levels were lowest in GCIMP positive or IDH1 mutant tumors, good prognostic indicators. However, Glut3 remained informative for poor prognosis in Proneural GCIMP negative or IDH1 wildtype tumors (Supplementary Fig. 6). These data further implicate Glut3 as an important regulator of GBM patient survival.
Tumor microenvironments, including hypoxia, are negative prognostic indicators of therapeutic response and patient survival. In the glioma TCGA dataset, high levels of a hypoxia marker, carbonic anhydrase IX (CA9)33, correlated with poor patient survival (Supplementary Fig. 7a). As Glut3 and Glut1 are reported to be hypoxia response genes, we determined whether the contribution of Glut3 to patient survival was a surrogate for hypoxia. While Glut3 (Supplementary Fig. 7b) and Glut1 (Supplementary Fig. 7c) correlated with CA9 expression, Glut3 (Supplementary Fig. 8) but not Glut1 (Supplementary Fig. 9) was informative for patient survival independent of hypoxia. Collectively, these data support a unique role of Glut3 in tumor growth and the malignancy of gliomas.
Glut3 Correlates with Pluripotency and Survival in Many Cancers
Glut3 is strongly associated with BTIC function and patient outcome. We, therefore, interrogated Glut3 in relation to a stem-cell state and patient survival in other solid tumors that follow a TIC model. As noted above, core stem cell regulators – Nanog, Sox2, Oct4 – are expressed by BTICs and are induced upon nutrient restriction during which Glut3 is critical. Thus, the role of Glut3 in BTICs could be a phenocopy of roles in pluripotency and reprogramming. Indeed, analysis of induced pluripotency datasets demonstrated that embryonic stem cells or induced pluripotent stem cells (iPS) express markedly elevated levels of Glut3 in comparison to fibroblasts34,35 (Fig. 8a, 8b and Supplementary Fig. 10d).
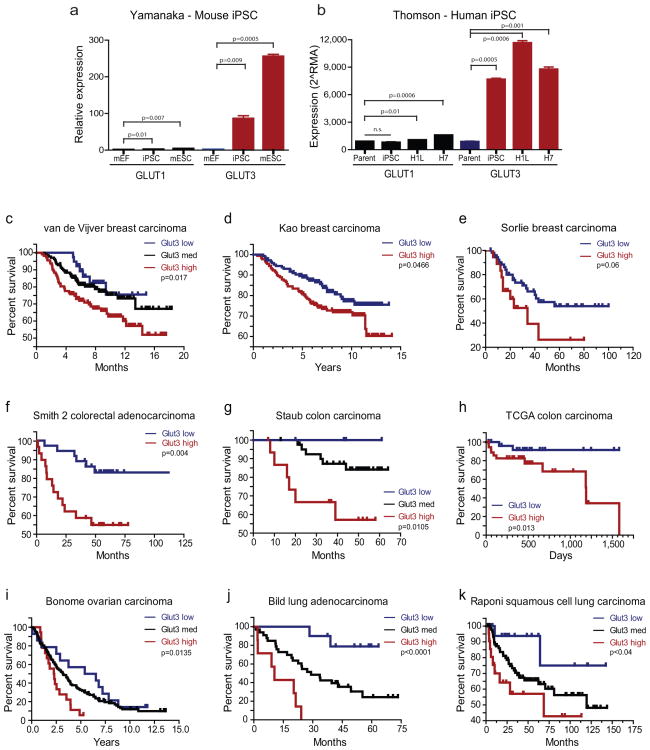
Figure 8. Glut3 Expression Correlates with Induced Pluripotency and Predicts Poor Survival in Multiple Tumor Types Beyond the Brain.
(a) Analysis of the Yamanaka dataset for induced pluripotent stem cell (iPSC) generation from mouse embryonic fibroblasts (mEF) demonstrates a significant increase in Glut3 expression with reprogramming. Mouse embryonic stem cells (mESC) also express high levels of Glut3 relative to fibroblasts [*, p=0.01; **p<0.01 with ANOVA comparison to mouse embryonic fibroblasts]. (b) Analysis of the Thomson dataset for iPSC generation from human foreskin fibroblasts (parent) demonstrates increases in Glut3 levels with reprogramming. Human embryonic stem cell lines (H1L and H7) also express high levels of Glut3 relative to fibroblasts [*, p=0.01; ** p<0.01 with ANOVA comparison to parental cells]. (c-k) Analysis of Carcinoma Datasets available through Oncomine indicates a significant correlation between high Glut3 expression and poor survival for breast (c-e), colorectal (f-h), ovarian (i) and lung (j-k) cancers. (c) van de Vijver Breast Carcinoma Dataset [n=39 for Glut3 low; n=138 for Glut3 medium; n=112 for Glut3 high; p=0.0107 with log-rank analysis]. (d) Kao Breast Carcinoma dataset [n=145 Glut3 low; n=181 Glut3 high; p=0.0466 with log-rank analysis]. (e) Sorlie breast carcinoma dataset [n=54 Glut3 low; n=18 Glut3 high; p=0.06 with log-rank analysis]. (f) Smith 2 Colorectal Adenocarcinoma Dataset [n=9 Glut3 low; n=34 Glut3 medium; n=11 Glut3 high; p=0.004 with log-rank analysis]. (g) Staub Colon Carcinoma dataset [n=4 Glut3 low; n=42 Glut3 medium; n=16 Glut3 high; p=0.0105 with log-rank analysis]. (h) TCGA colon carcinoma dataset [n=85 Glut3 low; n=68 Glut3 high; p=0.013 with log-rank analysis]. (i) Bonome Ovarian Carcinoma Dataset [n=24 Glut3 low; n=117 Glut3 medium; n=18 Glut3 high; p=0.0135 with log-rank analysis]. (j) Bild Lung Adenocarcinoma Dataset [n=10 Glut3 low; n=33 Glut3 medium; n=7 Glut3 high; p<0.0001 with log-rank analysis]. (k) Raponi Squamous Cell Lung Carcinoma Dataset [n=18 Glut3 low; n=90 Glut3 medium; n=21 Glut3 high; p<0.04 with log-rank analysis].
Glut3 could have a role in patient outcomes in TIC containing cancers beyond the brain. Prior reports have suggested Glut3 can be expressed in some cancers36-39, but Glut3 biological function and significance for patient outcomes is largely unknown with Glut3 association with poor survival reported in lung37, oral squamous38 and laryngeal39 carcinomas. Using Oncomine, we found that Glut3 expression correlated with poor survival in a broad range of tumor types, including those of the breast (Figs. 8c-e), colon (Figs. 8f-h), ovary (Fig. 8i), and lung (Figs 8j-k). These data suggest that many cancers dysregulate Glut3 to drive tumor growth and indicate a potential role for Glut3 in the TIC fraction of these tumors. The data also suggest that targeting of Glut3 could represent an important anti-cancer strategy.
Discussion
Metabolic reprogramming is a hallmark of cancer required for cellular transformation. Cancer cells are generally highly metabolically active, requiring both a constant source of energy and carbon for anabolic processes such as cell division and growth factor production. These needs are met through a shift to anaerobic glycolysis, which provides a cancer cell with both energy and carbon sources in the form of pyruvate. However, the glucose demand of cancer cells is dramatically increased due to the usage of the glycolytic end products for processes other than energy generation. These observations are challenging to resolve in light of the frequent impaired nutrient availability for cancers. Our current results suggest that the stem-like cells in malignant primary brain tumors co-opt high affinity transporter systems used by neurons to maintain access to available nutrients even in scarcity. The neuronal glucose transporter Glut3 is a high affinity glucose transporter (Km is approximately one-fifth of that of Glut1) permitting neurons to outcompete for nutrients where environmental concentrations are low, preserving their function and survival40. Our data demonstrate that BTICs display elevated levels of Glut3 to permit preferential glucose uptake, suggesting BTICs have a competitive advantage particularly important in relatively harsh microenvironments. More broadly, BTICs have differential molecular profiles to permit metabolic adaptations influencing GBM cell behaviors and maintaining tumor growth.
Exploiting the difference in glucose uptake in normal and neoplastic tissues with PET imaging permits non-invasive diagnosis or monitoring of tumor progression. These clinical observations in combination with our results indicating increased glucose uptake in BTICs suggest that PET imaging could be utilized to visualize the BTIC fraction. However, glucose metabolic rate and standardized uptake value did not significantly correlate with Glut3 expression in malignant gliomas41 and correlations with Glut1 expression have been mixed41,42. Data suggest that glucose metabolic rate correlates most strongly with cell proliferation rather than Glut expression in glioma42. As we have determined a relative difference in glucose uptake with preferential, but not exclusive, uptake in BTICs as well as Glut expression in both BTICs and non-BTICs, it is unlikely that any changes in PET imaging would be sufficient to define a TIC compartment.
Metabolism has been poorly linked to cancer cellular hierarchies, but metabolic changes are known to drive epigenetic modifications which influence cell fate. For example, mutations in IDH143-44 in low grade astrocytomas and secondary GBMs (but not primary GBMs predominately studied here) result in production of a new metabolic end product (2-hydroxyglutarate) leading to hypermethylation of DNA and repression of differentiation32,45. Targeting mutant IDH1 activity induces cellular differentiation and reduces tumor growth46, further supporting the potential of anti-metabolic therapies as a strategy to target GBMs. While IDH1 mutation contributes to GBM development, patients with IDH1 mutation have a relatively longer median survival. As our data indicate that IDH1 mutation is associated with lower levels of Glut3, it is likely that glucose metabolism differs in these tumors.
Although we have not fully explored the mechanisms regulating Glut3 upregulation in BTICs, the differences in Glut3 expression with GCIMP status indicate the possibility that aberrant methylation of Glut3 could be an important mechanism controlling its expression in cancer. Recent publications indicate a role for DNA methylation in the regulation of Glut3 expression in neurons and placenta47,48. As cancers display a stem cell epigenetic signature49,50, critical roles of metabolism in influencing epigenetics likely impact TIC phenotypes. Indeed, induced pluripotent stem cells express markedly elevated levels of Glut3 in comparison to parental fibroblasts, suggesting that upregulation of Glut3 is directly associated with acquisition of a stem cell state.
Cancer cell pro-tumorigenic properties are modeled in cell culture, but ex vivo conditions used to propagate cell lines are quite different from those found in vivo. To identify novel cancer targets, we must therefore establish media formulations to better mimic tumor microenvironments. Once targets are identified, therapies should be directed towards those which drive tumor progression without being functionally required for non-neoplastic cells to reduce toxicity. The brain is a very metabolically demanding organ, but the increased glucose demand of cancer cells suggests there may be a therapeutic window for targeting Glut3. However, Glut3 is expressed in neurons throughout the normal brain, suggesting the potential for toxicity. Unlike cancer cells, neurons are post-mitotic, suggesting that anti-Glut3 combinatorial therapies with conventional drugs targeting rapidly proliferating cells may have effects on BTICs while sparing neurons, supporting a therapeutic index. In addition, the data correlating Glut3 with poor prognosis in tumor types outside the brain suggest that a Glut3 inhibitor that does not cross the blood-brain-barrier would be useful for treating many aggressive cancers with minimal toxicity. We anticipate that understanding how TICs can outcompete for critical resources to maintain or promote tumor growth will represent a significant advancement in our understanding of malignant tumors including GBM and our ability to treat this devastating disease.
Methods
Cell Isolation and Culture
Glioblastoma (GBM) cells were derived from specimens of neurosurgical resection directly from patients in accordance with a Duke University or Cleveland Clinic Institutional Review Board-approved protocol in which informed consent was obtained by the tumor bank which provided deidentified excess tissue to the laboratory. Unenriched cells, BTICs, and/or non-BTICs were separated from GBM surgical specimens or xenografts as previously described5,12. Due to concerns about selection of cells in culture resulting in upregulation of Glut3 (Figure S2c,d), all cells were utilized within 5 tissue culture passages following dissociation. The cancer stem cell phenotype of CD133+ cells was confirmed by functional assays of self-renewal, stem cell marker expression, and tumor propagation. The CD133-depleted cells did not share these properties and were used in matched assays as non-BTICs. CD133-enriched BTICs were cultured in Neurobasal media with B27 (without vitamin A, Invitrogen), basic fibroblast growth factor (20 ng/ml) and epidermal growth factor (20 ng/ml), on petri dishes. CD133-depleted non-BTICs were cultured in Dulbecco's modified Eagle media (DMEM) with 10% fetal bovine serum, on tissue culture coated plates. Unenriched populations were cultured in BTIC media with 1% fetal bovine serum added, on tissue culture coated plates. The glucose concentration of all standard media formulations used was 4.5 g/L. For experiments with standard and restricted glucose, a restricted glucose environment was produced through a combination of one part standard media to nine parts glucose-free media, otherwise supplemented identically, resulting in a final media glucose concentration one tenth of standard media glucose, at 0.45 g/L. Due to poor viability of trypsin-EDTA treated glucose restricted cultures, all cultures were passaged using mechanical disaggregation and lifted from tissue culture plates.
Brain Tumor Specimen Characteristics
Based on microarray analysis, utilized specimens were subtyped as follows: Proneural (3832, IN528), Classical (4121, GBM43, GBM12, PB1), and Mesenchymal (IN326).
Quantitative RT-PCR
Total cellular RNA was isolated with the RNeasy kit (Qiagen) and reverse transcribed into cDNA using the Superscript III Reverse Transcription Kit (Invitrogen). Real time PCR was performed on an Applied Biosystems 7900HT cycler using SYBR-Green Mastermix (SA Biosciences) and gene-specific primers as follows: β-actin forward 5′-AGA AAA TCT GGC ACC ACA CC-3′ and reverse 5′-AGA GGC GTA CAG GGA TAG CA-3′; Oct4 5′-tctcccatgcattcaaactgag-3′ and reverse 5′- cctttgtgttcccaattccttc-3′; NANOG forward 5′-gaaatacctcagcctccagc-3′ and reverse 5′-gcgtcacaccattgctattc-3; Sox2 forward 5′- cacactgcccctctcac-3′ and reverse 5′-tccatgctgtttcttactctcc-3′; Glut1 forward 5′-ATCGTGGCCATCTTTGGCTTTGTG-3′ and reverse 5′-CTGGAAGCACATGCCCACAATGAA-3′; Glut2 forward 5′-AGCTGCATTCAGCAATTGGACCTG-3′ and reverse 5′-ATGTGAACAGGGTAAAGGCCAGGA-3′; Glut3 forward 5′-AGCTCTCTGGGATCAATGCTGTGT-3′ and reverse 5′-ATGGTGGCATAGATGGGCTCTTGA-3′; Glut4 forward 5′-TCGTGGCCATATTTGGCTTTGTGG-3′ and reverse 5′-TAAGGACCCATAGCATCCGCAACA-3′
Flow Cytometry Glut surface staining
For FACS staining of surface Glut expression, live cells were incubated with either Glut3 (R&D, MAB1415) or Glut1 (Abcam AB40084), and CD133 (AC133-APC, Miltenyi) for 45 min at dilutions specified in the manufacturer's protocols.
In vitro Limiting Dilution Neurosphere Formation Assay
For in vitro limiting dilution assays, propidium iodide negative cells were sorted by FACS with decreasing numbers of cells per well (50, 20, 10, 5, and 1) plated in 96 well plates containing Neurobasal media with B27 (without vitamin A, Invitrogen), basic fibroblast growth factor (20 ng/ml) and epidermal growth factor (20 ng/ml). Extreme limiting dilution analysis was performed using software available at http://bioinf.wehi.edu.au/software/elda. Neurosphere formation assays were also performed similar to our prior report12 with 10 cells per well plated in 96 well plates and the percent of wells with neurospheres measured after 10 days.
Competition Mixing Assay
Unsorted glioma cells were infected with either RFP or GFP lentivirus. Following infection, cells were sorted for color, viability using LIVE/DEAD Fixable Blue Dead Cell Stain kit (Invitrogen) and CD133-APC status by flow cytometry and directly plated in Neurobasal stem cell media with 1% FBS on Geltrex-coated plates. Cells were imaged on days 4 and 7, and collected for flow cytometry analysis on day 7.
Cell Death Assay
Apoptosis and Necrosis Quantitation kit was obtained from Biotium and performed according to the manufacturer's protocol, with cell fixation following staining.
pGZ-Nanog non-BTIC Adaptation Imaging
pGreenZeo-Nanog plasmid were obtained from System Biosciences, prepared into lentiviral particles, and used to infect cells. Infected cells were sorted via flow cytometry to obtain only GFP expressing cells. Fourteen days post-sort, cells were plated on Geltrex in Neurobasal stem cell media with 1% FBS and either full or restricted glucose. Twenty four hours post-plating, images were taken on a Leica DMIRB Inverted Microscope equipped for time-lapse microscopy with a Roper Scientific CoolSNAP HQ Cooled CCD camera (Roper Scientific, Tucson AZ, USA), temperature controller (37°C) and CO2 (5%) incubation chamber (Leica Microsystems GmbH), PeCon incubator (PeCon GmbH, Erbach, Germany), Prior motorized stage with linearly encoded controller with x/y/z drive for time-lapse imaging of multiple fields and heating insert for 6-well plates (Prior Scientific Inc., Rockland, MA, USA), Uniblitz shutter (Vincent Associates, Rochester, NY, USA), and MetaMorph Software (Molecular Devices, Downingtown, PA, USA). Bright field images were taken every ten minutes and green fluorescence images were taken every twenty minutes for 72 hours.
Low Glucose non-BTIC Adaptation Neurosphere Formation Assay
For the demonstration of non-BTIC adaptation to low glucose, a slightly modified version of the above neurosphere formation assay was employed. CD133 negative, propidium iodide negative cells were sorted in decreasing numbers of cells (as above) into 96 well plates containing either stem cell media or restricted glucose stem cell media. Cells were left in this media for seven days, at which point glucose was added to all wells to eliminate any possible effect of glucose restriction on proliferation, which would affect the sphere formation results. Fourteen days after glucose addition (twenty one days post plating), spheres were counted as above. The frequency of adaptive non-BTICs was calculated as the difference in sphere formation between the normal glucose condition and the 7 days restricted glucose condition.
Non-Fluorescent Glucose Uptake Assay
Following 45 minutes of glucose starvation, BTIC or non-BTIC cells were incubated in 0.45 g/L glucose for 30 minutes, then collected for lysis via mechanical lifting. Cells were lysed in RIPA buffer (Sigma) and the protein was quantified as a loading control via Bradford assay. A glucose colorimetric detection kit was obtained (Arbor Bioassays) and performed according to protocol, with the exception of an increased incubation time to account for the use of whole cell lysate.
Fluorescent Glucose Uptake Assay
Following 30 minutes of glucose starvation, GBM cells were incubated for 30 minutes in the presence of a fluorescent glucose analog, 2-[N-(7-nitrobenz-2-oxa-1,3-diaxol-4-yl)amino]-2-deoxyglucose (2-NBDG). For studies comparing uptake in CD133+ and CD133- cells, unenriched cells were co-incubated with AC133-APC antibody and analyzed via FACS. For studies with shRNA expressing BTICs, cells were washed and fluorescence analyzed using a VICTOR plate reader.
Ex vivo Glucose Uptake Imaging Assay
Prior to transplantation onto brain slices for imaging, BTICs were labeled with Cell Tracker Red CMPTX (Invitrogen) and non-BTICs were labeled with Cell trace far red DDAO-SE (Invitrogen). For ex vivo glucose analysis, slice cultures were prepared from mice according to prior publications51 and 5000 cells total were transplanted (at a ratio of 1:1, BTIC:non-BTIC). Transplanted cells were incubated overnight to ensure integration and survival in the brain slices and prior to imaging, slices were incubated in 50μg/ml NDBG for 30 minutes prior to image acquisition. Imaging was done using a Leica multiphoton microscope as previously described52 with a 20× liquid immersion objective, NA=1.0. Images were acquired at 820nm and processed using Imaris software (Bitplane).
Immunofluorescent Staining
For immunostaining analysis at the single cell level, live cells isolated via a ficoll gradient were collected on the Cell-Tack (BD) coated coverslips by centrifugation and fixed with 4% formaldehyde for 15 min followed by post-fixed/permeabilized with cold methanol for 20 min. Cells were blocked in 5% goat serum for 30 minutes followed by incubation with Glut3 (R&D MAB1415) and Sox2 (MAB2018) primary antibodies at manufacturer recommended dilutions overnight at 4°C followed by the appropriate isotype-specific secondary fluorescently labeled antibodies (Invitrogen Molecular Probes) for 1 hour at room temperature. Nuclei were counterstained with Hoechst 33342. Wide field images were taken using a Leica fluorescent microscope.
Vectors and Lentiviral Transfection
Lentiviral clones expressing SLC2A3 shRNAs (TRC0000043616 and TRC0000043615) and control shRNA (SHC002) were purchased from Sigma-Aldrich. Viral particles were produced in 293T cells with the pPACK set of helper plasmids (System Biosciences) in stem cell medium. Glut3 shRNA1: designed against the coding sequence 5′-CCGGCTTGGTCTTTGTAGCCTTCTTCTCGAGAAGAAGGCTACAAAGACCAAGTTTTTG-3′; Glut3 shRNA2: designed against the coding sequence 5′- CCGGAGTAGCTAAGTCGGTTGAAATCTCGAGATTTCAACCGACTTAGCTACTTTTTTG-3′
In vivo Tumor Initiation Assay
All animal procedures were performed in accordance with Cleveland Clinic IACUC approved protocols. Animals were housed in a temperature-controlled vivarium with a 14 hour light, 10 hour dark cycle at no more than 5 animals per cage. For nutrient deprivation studies, unenriched GBM cells were cultured in standard or low glucose conditions for seven days and 5,000 viable cells were intracranially injected into athymic/nude mice as previously described12. For shRNA studies, BTICs were transduced with lentiviral vectors expressing Glut3 shRNA or non-targeting control shRNA and 1000 viable cells were intracranially injected into female athymic/nude mice between 4 and 6 weeks of age. Animals were maintained until development of neurological signs (e.g. lethargy, ataxia, seizures and/or paralysis) when brains were harvested. If no neurologic signs developed within 105 days, animals were subsequently sacrificed and the experiment terminated. Harvested brains were fixed in 4% formaldehyde, sunk in 30% sucrose, cryopreserved in OCT, and cryosectioned.
Retrospective analysis of Glut3 gene expression in human gliomas
Correlations between glioma grade, patient survival, tumor recurrence and Glut3 gene expression were determined through analysis of TCGA, Sun, Nutt, Freije, and Philips brain datasets respectively which are available through Oncomine (Compendia Biosciences, www.oncomine.org). High and low groups were defined as above and below the mean respectively. For analysis with high, medium, and low groups, high was defined as greater than one standard deviation above the mean, low is greater than one standard deviation below the mean, and medium is within one standard deviation of the mean. The National Cancer Institute's Repository for Molecular Brain Neoplasia Data (REMBRANDT, http://rembrandt.nci.nih.gov) was also evaluated for correlations between glioma patient survival and gene expression with up- or downregulation being defined as a 2 fold change relative to mean values.
Statistical analysis
All grouped data are presented as mean ± standard error. Difference between groups was assessed by ANOVA or Student's t-test using GraphPad Prism software. For survival analysis, Kaplan Meier curves were generated using either MedCalc or Prism software and log rank analysis performed. All experiments were repeated in each specimen presented in at least duplicate with triplicate technical replicates. We utilize short term passaged cells and numbers of BTICs isolated form any one specimen or xenograft are low (usually 10% or less of the bulk tumor cells). Therefore, we have not repeated all experiments with every sample utilized in the entire manuscript. We used isolated cells from different specimens to address specific questions in separate replicated experiments. We observed consistent results across specimens when similar experiments were performed as shown. Data distribution was assumed to be normal but this was not formally tested. Data and animals were not randomized but were collected and processed in appropriate experimental arms. Data collection and analysis were not performed blind to the conditions of the experiments.
Sample sizes
For mouse experiments, sample sizes were calculated using the formula n=1+2C(s/d)ˆ2 where: n=number of animals per arm, C=7.85 (significance level of 5% with a power of 80%), s = standard deviation, and d = difference to be detected. For other experiments, no statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those in our previous publications.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health: CA129958, CA116659, and CA154130 (J.N.R.); CA151522 (A.B.H); CA157948 (J.D.L.), CA137443, NS063971, CA128269, CA101954, and CA116257 (A.E.S); James S. McDonnell Foundation (J.N.R.), Voices Against Brain Cancer (J.D.L), Ohio Department of Development Tech 09-071 (A.E.S.), American Brain Tumor Association (Y.K.); Melvin Burkhardt Chair in Neurosurgical Oncology and Karen Colina Wilson Research Endowment (R.J.W.); Cleveland Clinic Foundation Tissue Procurement Service; S. Staugatis; M. McGraw; Flow Cytometry Core; Shideng Bao, Tyler Miller, David Schonberg, and Monica Venere.
Footnotes
The authors have no financial conflicts of interest that may influence the results or data interpretation.
Competing Interests Statement: We have no competing interests.
Supplemental Information: Supplemental information includes ten figures, four tables, one video, and methods which can be found with this article online.
Author Contributions: Conception and experimental design: William Flavahan, Qiulian Wu, Youngmi Kim, Justin Lathia, Jeremy N. Rich, Anita B. Hjelmeland
Methodology and data acquisition: William Flavahan, Qiulian Wu, Masahiro Hitomi, Nasiha Rahim, Andrew E. Sloan, Robert J. Weil, Ichiro Nakano, Jann Sarkaria, Brett W. Stringer, Meizhang Li, Justin D. Lathia, Anita B. Hjelmeland
Analysis and interpretation of data: William Flavahan, Masahiro Hitomi, Meizhang Li, Justin D. Lathia, Jeremy N. Rich, Anita B. Hjelmeland
Manuscript writing and/or revision: William Flavahan, Jeremy N. Rich, Anita B. Hjelmeland
References
- 1.Stupp R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 3.Galli R, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 4.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 5.Bao S, et al. Brain Tumor Initiating Cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 6.Liu G, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2005;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters A, et al. The selfish brain: Competition for energy resources. J Gen Physiol. 2004;28:143–180. doi: 10.1016/j.neubiorev.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 9.Derr RL, et al. Association between hyperglycemia and survival in patients with newly diagnosed glioblastoma. J Clin Oncol. 2009;27:1082–1086. doi: 10.1200/JCO.2008.19.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panopoulos AD, et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, et al. Hypoxia-inducible factors regulate tumorigenic capacity of Brain Tumor Initiating Cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8:3274–3284. doi: 10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hjelmeland AB, et al. Acidic stress promotes a Brain Tumor Initiating Cell phenotype. Cell Death Differ. 2011;18:829–840. doi: 10.1038/cdd.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fellows LK, Boutelle MG. Rapid changes in extracellular glucose levels and blood flow in the striatum of the freely moving rat. Brain Res. 1993;604:225–231. doi: 10.1016/0006-8993(93)90373-u. [DOI] [PubMed] [Google Scholar]
- 16.Burgess EA, Sylven B. Glucose, lactate, lactic dehydrogenase activity in normal interstitial fluid and that of solid mouse tumors. Cancer Res. 1962;22:581–588. [PubMed] [Google Scholar]
- 17.Laks DR, et al. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells. 2009;27:980–987. doi: 10.1002/stem.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathews EH, Liebenberg L, Pelzer R. High-glycolytic cancers and their interplay with the body's glucose demand and supply cycle. Med Hypotheses. 2011;76:157–165. doi: 10.1016/j.mehy.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Yoshioka K, et al. A novel fluorescent derivative of glucose applicable to the assessment of glucose uptake activity of Escherichia coli. Biochim Biophys Acta. 1996;1289:5–9. doi: 10.1016/0304-4165(95)00153-0. [DOI] [PubMed] [Google Scholar]
- 20.Song J, et al. Neuronal circuitry mechanism regulating adult quiescent neural stem-cell fate decision. Nature. 2013;489:150–154. doi: 10.1038/nature11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vannucci SJ, Maher F, Simpson IA. Glucose transporter proteins in brain: delivery of glucose to neurons and glia. Glia. 1997;21:2–21. doi: 10.1002/(sici)1098-1136(199709)21:1<2::aid-glia2>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 22.Nagamatsu S, Sawa H, Wakizaka A, Hoshino T. Expression of facilitative glucose transporter isoforms in human brain tumors. J Neurochem. 1993;61:2048–2053. doi: 10.1111/j.1471-4159.1993.tb07441.x. [DOI] [PubMed] [Google Scholar]
- 23.Boado RJ, Black KL, Pardridge WM. Gene expression of Glut3 and Glut1 glucose transporters in human brain tumors. Brain Res Mol Brain Res. 1994;27:51–57. doi: 10.1016/0169-328x(94)90183-x. [DOI] [PubMed] [Google Scholar]
- 24.Freije WA, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 25.Sun L, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 27.Nutt CL, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- 28.Madhavan S, et al. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7:157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network. Comprehensive genomic characterization define human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loncaster JA, et al. Carbonic anhydrase (CA IX) expression, a potential new intrinsic marker of hypoxia: correlations with tumor oxygen measurements and prognosis in locally advanced carcinoma of the cervix. Cancer Res. 2001;61:6394–6399. [PubMed] [Google Scholar]
- 34.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 35.Yu J, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Younes M, Lechago LV, Somoano JR, Mosharaf M, Lechago J. Immunohistochemical detection of Glut3 in human tumors and normal tissues. Anticancer Res. 1997;17:2747–2750. [PubMed] [Google Scholar]
- 37.Younes M, Brown RW, Stephenson M, Gondo M, Cagle PT. Overexpression of Glut1 and Glut3 in stage I nonsmall cell lung carcinoma is associated with poor survival. Cancer. 1997;80:1046–51. doi: 10.1002/(sici)1097-0142(19970915)80:6<1046::aid-cncr6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 38.Ayala FR, et al. Glut1 and Glut3 as potential prognostic markers for Oral Squamous Cell Carcinoma. Molecules. 2010;15:2374–2387. doi: 10.3390/molecules15042374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baer S, Casaubon L, Schwartz MR, Marcogliese A, Younes M. Glut3 expression in biopsy specimens of laryngeal carcinoma is associated with poor survival. Laryngoscope. 2002;112:393–396. doi: 10.1097/00005537-200202000-00034. [DOI] [PubMed] [Google Scholar]
- 40.Gould GW, Holman GD. The glucose transporter family: structure, function and tissue-specific expression. Biochem J. 1993;295:329–341. doi: 10.1042/bj2950329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charnley N, et al. No relationship between 18F-fluorodeoxyglucose positron emission tomography and expression of Glut-1 and -3 and hexokinase I and II in high-grade glioma. Oncol Rep. 2008 Sep;20(3):537–42. [PubMed] [Google Scholar]
- 42.Chung JK, et al. Comparison of [18F]fluorodeoxyglucose uptake with glucose transporter-1 expression and proliferation rate in human glioma and non-small-cell lung cancer. Nucl Med Commun. 2004 Jan;25(1):11–7. doi: 10.1097/00006231-200401000-00003. [DOI] [PubMed] [Google Scholar]
- 43.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohle D, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novakovic B, Gordon L, Robinson WP, Desoye G, Saffery R. Glucose as a fetal nutrient: dynamic regulation of several glucose transporter genes by DNA methylation in the human placenta across gestation. J Nutr Biochem. 2013;24:282–288. doi: 10.1016/j.jnutbio.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, Shin BC, Thamotharan S, Devaskar SU. Creb1-Mecp2-(m)CpG complex transactivates postnatal murine neuronal glucose transporter isoform 3 expression. Endocrinology. 2013;d154611:1598. doi: 10.1210/en.2012-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Widschwendter M, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 50.Wong DJ, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 52.Lathia JD, et al. Direct in vivo evidence for tumor propagation by glioblastoma cancer stem cells. PLoS One. 2011;6:e24807. doi: 10.1371/journal.pone.0024807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.