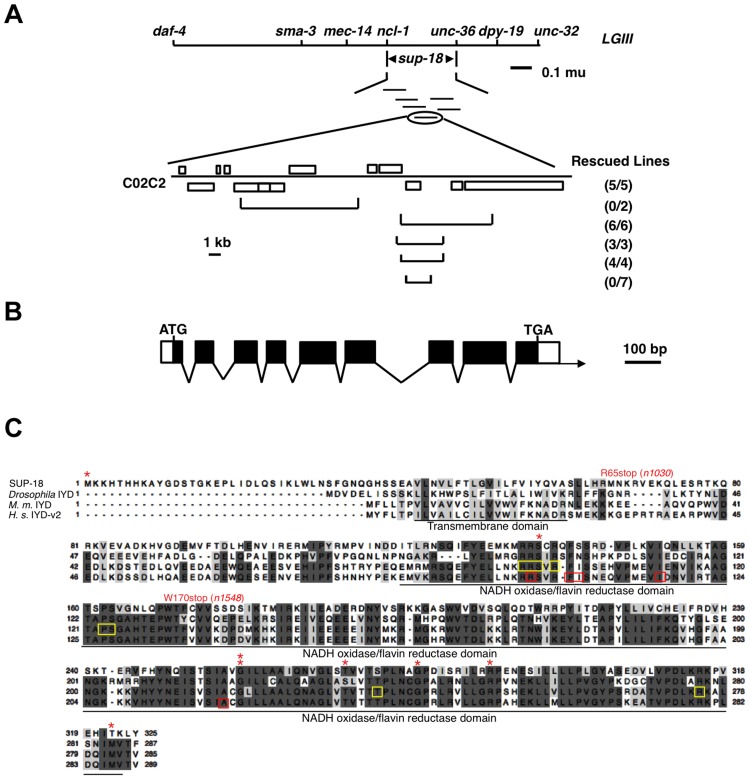
Figure 1. sup-18 encodes the C. elegans ortholog of mammalian iodotyrosine deiodinase.
(A) Top, genetic map of the sup-18 region of linkage group III (LG III). Horizontal lines below sup-18 represent the cosmids tested for rescue of sup-18(n1010); sup-10(n983gf) mutants. Bottom, physical map of cosmid C02C2. Open boxes, coding regions; horizontal brackets, cosmid subclones. Rescued lines, number of independently derived transgenic lines/total number of lines scored. Rescue was scored as the appearance of animals with the phenotype of rubberband Unc paralysis. (B) Intron-exon structure of sup-18 as inferred by comparison of the cDNA and genomic sequences. Dark boxes, coding regions; open boxes, untranslated regions; arrow, direction of transcription. The sup-18 open reading frame is 978 bp, the 5′ UTR is 57 bp and the 3′UTR is 44 bp. (C) Sequence alignment of SUP-18 and iodotyrosine deiodinases from other species. Amino acids conserved in at least three species are darkly shaded, while amino acids with similar physical properties in at least three species are lightly colored. *: sup-18 missense mutations (see Table 3). Residues mutated in human hypothyroidism patients are indicated by red boxes. The FMN cofactor binding residues in mouse IYD are indicated by yellow boxes. The transmembrane domain and NADH oxidase/flavin reductase domain are underlined and labeled. Genebank accession numbers are as follows: SUP-18, JX978835; Drosophila, AAM11009; M. musculus, AAH23358; H. sapiens, NP_981932.