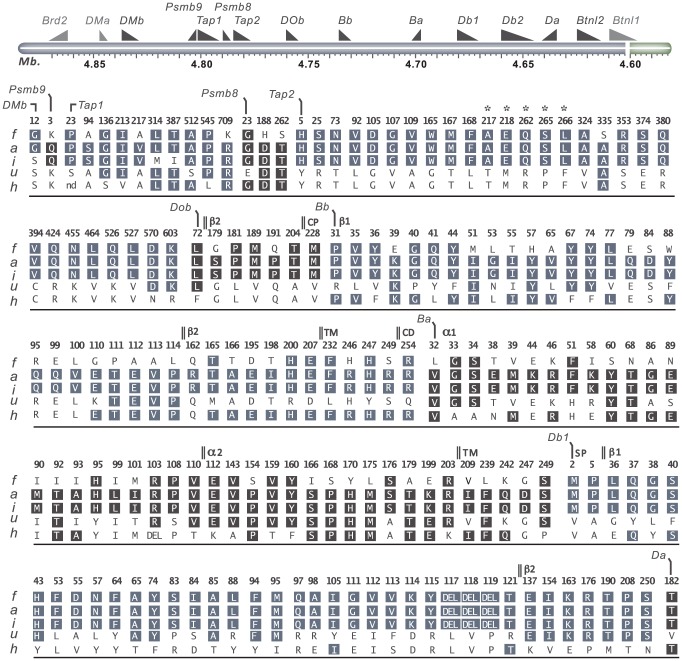
Figure 4. Coding variants in T cell selection QTL 2 (Tcs2).
Nonsynonymous- and structural variants in Tcs2 were determined by Sanger sequencing of DA.1F (f), DA (a), DA.1I (i), DA.1U (u) and DA.1H (h) (see also Table 2). Amino acid substitutions are indicated in standard single letter codes and insertions/deletions as DEL. Gene annotations are from UniProt; protein domains depicted in DOb are derived from the human homolog. Letters in boxes depict residues with background allele (DA) with numbers above indicating amino acid positions in the translated cds. On top is a schematic illustration of the genes in the region according to the 3.4 genome assembly (genes outside the QTL are shown in gray). Annotations used: TM, transmembrane domain; CD, cytoplasmic domain; CP, connecting peptide; β1, beta 1 domain; β2, beta 2 domain; α1, alpha 1 domain; α2, alpha 2 domain. *Residues associated with class-I modification (cim).