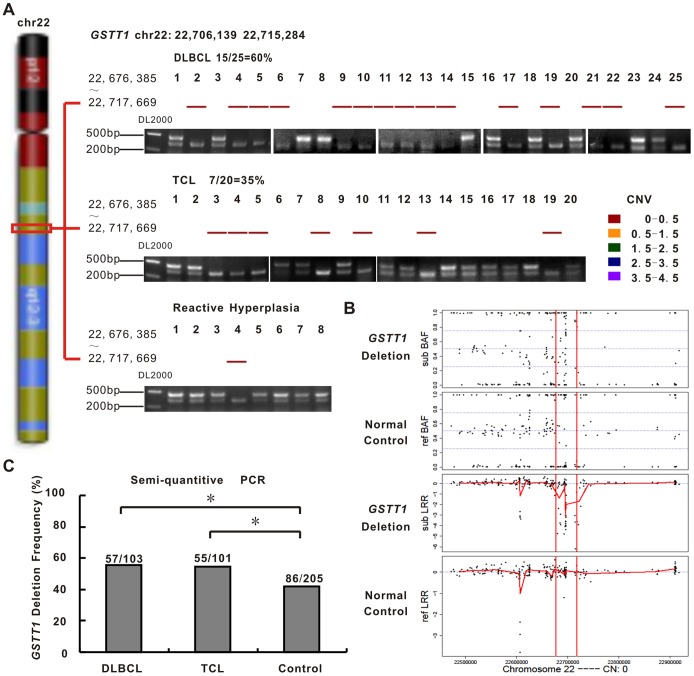
Figure 1. GSTT1-null genotype is frequently observed in patients with lymphoma.
A: GSTT1 deletion detected by copy number variation (CNV) analysis (upper panel) and validated by semi-quantitative PCR (lower panel) in diffuse large B-cell lymphoma (DLBCL, 15/25, 60.0%), T-cell lymphoma (TCL, 7/20, 35.0%) and reactive hyperplasia (1/8, 12.5%). B: SNP data illustrating the distribution of B allele frequencies (BAF) and Log R Ratio (LRR) values across the region of chromosome 22 (22,676,385 bp to 22,717,669 bp) in the GSTT1-deleting cases and normal controls. C: Semi-quantitative PCR validation of GSTT1 deletion in expanded sample-set of DLBCL (57/103, 55.3%), TCL (55/101, 54.5%) and normal controls (86/205, 42.0%). *P<0.05 comparing with normal controls.