Abstract
Background
Emerging evidence indicated that common polymorphisms of NOD2 might impact individual susceptibility to cancer. However, the results from published studies were inconclusive. The aim of this meta-analysis was to elucidate whether NOD2 polymorphisms were associated with cancer risk.
Methods
A systematically literature search was performed by using electronic databases including PubMed and Web of Science. ORs and their 95% CI were used to assess the strength of association between NOD2 gene polymorphisms and cancer risks.
Results
Thirty case-control studies were included in this meta-analysis. The pooled analysis indicated that NOD2 rs2066842 C/T polymorphism was not significantly associated with cancer risk; for NOD2 rs2066844 C/T polymorphism, (TT+CT) genotype was associated with increased cancer risk compared with wild-type CC genotype (OR = 1.32, 95% CI = 1.01–1.72, P = 0.041); for NOD2 rs2066845 C/G polymorphism, individuals with (CC+CG) genotype were significantly associated with increased cancer risk compared with GG genotype (OR = 1.32, 95% CI = 1.01–1.72, P = 0.040); for NOD2 rs2066847 (3020insC) polymorphism, carriers of (insC/insC+insC/−) genotype were significantly associated with increased cancer risk compared with −/− carriers (OR = 1.23, 95% CI = 1.10–1.38, P<0.001). In the subgroup analysis of cancer type, (insC/insC+insC/−) genotype was significantly associated with increased risk of colorectal cancer, gastric cancer and MALT lymphoma, breast cancer, lung cancer, laryngeal cancer but not with urogenital cancer, pancreatic cancer, melanoma or non-Hodgkin lymphoma.
Conclusion
NOD2 rs2066844 C/T, rs2066845 C/G and rs2066847 (3020insC) polymorphisms might be associated with increased cancer risk. No significant association was observed between NOD2 rs2066842 C/T polymorphism and cancer risk. Further large-scale and well-designed studies are still needed to confirm the results of our meta-analysis.
Introduction
Cancer is a major health problem in the most parts of the world. Approximately 12.7 million cancer cases and 7.6 million cancer deaths are estimated to occur each year worldwide [1]. The prevention and treatment for cancers caused increasing financial burdens around the world [2]. As a complex disease, cancer is strongly influenced by environmental and genetic factors, of which gene polymorphism is a critical cause for the difference of individual genetic susceptibility to cancer [3]. Identification of the key gene polymorphisms that are associated with cancer risk is essential for predicting individual at risk.
The nucleotide-binding oligomerization domain containing 2 (NOD2) gene, also known as CARD15, is mapped to chromosome 16q21. NOD2 is a member of evolutionarily conserved Nod-like receptors (NLRs) family which share a tripartite structure of a C-terminal sensor domain (leucine-rich repeats, LRRs), a central nucleotide-binding oligomerization domain (NOD) and an N-terminal effector domain (CARD) [4]. NOD2 participates in sensing components of microbial cell wall and has been reported to regulate apoptosis and chronic inflammatory conditions [5]. The most commonly studied polymorphisms included three missense mutations (rs2066842 C/T, rs2066844 C/T, rs2066845 C/G) and a frameshift mutation (rs2066847 insC). These four polymorphisms were located at coding regions and might affect the expression and function of NOD2 by altering amino acid. Recently, increasing studies investigated the relations between these four polymorphisms and disease risk.
The rs2066842, rs2066844, rs2066845 and rs2066847 polymorphisms were initially found to be associated with increased risk of Crohn’s disease (CD) in 2001 to 2003 [6]. Subsequently, the relation between these polymorphisms and ulcerative colitis (UC) risk was revealed [7]. In 2004, Kurzawski et al. first linked NOD2 polymorphism to risk of colorectal cancer [8]. After that, increasing studies focused on the association between NOD2 polymorphisms and risks of various cancers including gastric cancer, colorectal cancer, endometrial cancer, breast cancer, ovarian cancer, laryngeal cancer and so on. However, the results from the individual studies were inconsistent.
So far, no comprehensive meta-analysis has investigated the overall cancer risk in relation to NOD2 polymorphisms, except for a meta-analysis only concerning colorectal cancer in 2010. To explore whether NOD2 polymorphisms were associated with risks of overall cancer and specific cancer subtypes, we performed a meta-analysis on the association between the four most frequently studied NOD2 polymorphisms (rs2066842 C/T, rs2066844 C/T, rs2066845 C/G and rs2066847 insC) and cancer risk in the present study.
Materials and Methods
Identification and Eligibility of Relevant Studies
Literatures of electronic databases including PubMed and Web of Science were systematically searched using the search terms of “NOD2/CARD15”, “polymorphism/mutation/variant” and “cancer/malignancy/neoplasm”. References cited in each identified literatures were further searched manually to find potential available studies. We contacted the author for specific raw data if the data presented in the article were not sufficient. When overlapping data exists, only the latest study with the largest sample was selected for this meta-analysis. The last search date was July 1, 2013.
Inclusion and Exclusion Criteria
Studies included in the present meta-analysis must meet the inclusion criteria as follows: observational studies concerning the association between NOD2 gene polymorphisms (rs2066842 C/T, rs2066844 C/T, rs2066845 C/G and rs2066847 insC) and cancer risks; studies published in English; studies with sufficient raw data for estimating odds ratios (OR) and their 95% confidence interval (CI); the control group of the studies should be in accordance with the Hardy Weinberg Equilibrium (HWE). The main reasons for exclusion were reviews or meta-analysis; animal experiments; not relevant to specific polymorphisms; duplicate publications; no raw data after contacting the author; studies not in English.
Data Extraction
Two authors (Jingwei Liu and Caiyun He) extracted the data from the included studies independently. The following information was extracted from each study: first author, year of publication, ethnicity of the population, numbers of cases and controls, detection methods of NOD2 polymorphism and the source of the control group. The conflicts were resolved after discussion and consensus was finally reached on all of the extracted data.
Statistical Analysis
The statistical analysis was performed by Stata software (Version 11.0; StataCorp, College Station, TX). ORs and their 95% CI were used to assess the strength of association between NOD2 gene polymorphisms and cancer risks. P value <0.05 was considered as statistically significant. Heterogeneity was measured by using Q statistic (P<0.10 indicates significant heterogeneity between studies) and I-squared (I2) value [9]. A fixed-effects model using Mantel-Haenszel method [10] was performed to calculate the pooled ORs when heterogeneity between studies was not significant. Otherwise, a random-effects model using DerSimonian and Laird method [11] was applied. Sensitivity analysis was performed to explore heterogeneity when significant heterogeneity was indicated. Subgroup analyses were performed to explore the effects of cancer type and source of controls. Additionally, publication bias were evaluated qualitatively by performing funnel plots and assessed quantitatively by Begg’s test [12] and Egger’s test [13], respectively. P value<0.05 for Begg’s and Egger’s tests indicates significant publication bias.
Results
Characteristics of the Included Studies
This meta-analysis was organized according to the PRISMA statement (Checklist S1). Totally 93 literatures were indentified through electronic databases after duplicates removal. After reviewing the titles and abstracts of the potential available articles, 57 records were excluded mainly because of no relevance, in vitro or animal experiments, reviews or meta-analysis. The left 36 full-text articles were further assessed for eligibility. Finally, 30 full-text articles with eligibility were included in this meta-analysis [8], [14]–[42]. The flow chart of article selection was presented in Figure S1.
The main characteristics of the studies included in this meta-analysis were summarized in Table 1. All the included studies were case-control designed published in English. The populations of the studies were all Caucasians. Four studies consisting of 368 cases and 567 controls investigated the association of NOD2 rs2066842 C/T (P268S) polymorphism and cancer risk; 16 studies including 4507 cases and 4780 controls studied the association of NOD2 rs2066844 C/T (R702W) polymorphism and cancer risk; 14 articles including 4185 cases and 4474 controls investigated the association of NOD2 rs2066845 C/G (G908R) polymorphism and cancer risk; for NOD2 rs2066847/rs5743293 (3020insC) polymorphism, 25 studies consisting of 23167 cases and 28601 controls were included. The types of cancers studied in relation to NOD2 polymorphisms included gastric cancer and MALT lymphoma, colorectal cancer (CRC), melanoma, endometrial cancer, pancreatic cancer, breast cancer, non-Hodgkin lymphoma, laryngeal cancer, lymphocytic leukaemia and ovarian cancer. Data concerning different cancers were treated as separate studies in the subgroup analysis.
Table 1. Characteristics of eligible studies in this meta-analysis.
| Author | Year | Ethnicity | Cancer type | Controls source | Case | Control | Genotyping method |
| rs2066842 C/T (P268S) | |||||||
| Roberts, R. L. | 2006 | New Zealander | Colorectal cancer | PB | 133 | 201 | ARMS |
| Wex, T. | 2008 | German | Gastric cancer | PB | 167 | 153 | PCR-RFLP |
| Szeliga, J. | 2008 | Polish | Rectal cancer | HB | 51 | 100 | PCR-RFLP |
| Hnatyszyn, A. | 2010 | Polish | Gastric cancer | HB | 17 | 113 | Pyrosequencing |
| rs2066844 C/T (R702W) | |||||||
| Debniak, T. | 2005 | Polish | Melanoma | HB | 470 | 649 | Allele-specific PCR |
| Papaconstantinou, I. | 2005 | Greek | Colorectal cancer | N.A. | 104 | 100 | Allele-specific PCR |
| Rosenstiel, P. | 2006 | German | Gastric MALT lymphoma | HB | 83 | 308 | Taqman |
| Roberts, R. L. | 2006 | New Zealander | Colorectal cancer | PB | 133 | 201 | ARMS |
| Lakatos, P. L. | 2007 | Hungarian | Colorectal cancer | N.A. | 194 | 200 | dHPLC |
| Vogel, U. | 2007 | Danish | Colorectal cancer | PB | 355 | 753 | CE-SSCP |
| Tuupanen, S. | 2007 | Finnish | Colorectal cancer | PB | 953 | 508 | ARMS |
| Szeliga, J. | 2008 | Polish | Rectal cancer | HB | 51 | 100 | PCR-RFLP |
| Suchy, J. | 2008 | Polish | Colorectal cancer | HB | 350 | 350 | PCR-RFLP |
| Ture-Ozdemir, F. | 2008 | Greek | Gastric MALT lymphoma | HB | 56 | 51 | PCR-RFLP |
| Wex, T. | 2008 | German | Gastric cancer | PB | 159 | 150 | PCR-RFLP |
| Mockelmann, N. | 2009 | German | Colorectal cancer | PB | 1044 | 724 | SNPlex |
| Angeletti, S. | 2009 | Italian | Gastric cancer | PB | 170 | 156 | PCR-ARMS |
| Freire, P. | 2010 | Portuguese | Colorectal cancer | PB | 112 | 152 | Real-time PCR |
| Rigoli, L. | 2010 | Italian | Gastric cancer | PB | 60 | 87 | PCR-RFLP |
| Ashton, K. A. | 2010 | Austrilian | Endometrial cancer | PB | 213 | 291 | Real-time PCR |
| rs2066845 C/G (G908R) | |||||||
| Papaconstantinou, I. | 2005 | Greek | Colorectal cancer | N.A. | 104 | 100 | PCR-RFLP |
| Debniak, T. | 2005 | Polish | Melanoma | HB | 470 | 649 | Allele-specific PCR |
| Rosenstiel, P. | 2006 | German | Gastric MALT lymphoma | HB | 83 | 308 | Taqman |
| Roberts, R. L. | 2006 | New Zealander | Colorectal cancer | PB | 133 | 201 | ARMS |
| Tuupanen, S. | 2007 | Finnish | Colorectal cancer | PB | 960 | 508 | ARMS |
| Lakatos, P. L. | 2007 | Hungarian | Colorectal cancer | N.A. | 194 | 200 | dHPLC |
| Vogel, U. | 2007 | Danish | Colorectal cancer | PB | 355 | 753 | CE-SSCP |
| Ture-Ozdemir, F. | 2008 | Greek | Gastric MALT lymphoma | HB | 56 | 51 | PCR-RFLP |
| Szeliga, J. | 2008 | Polish | Rectal cancer | HB | 51 | 100 | PCR-RFLP |
| Suchy, J. | 2008 | Polish | Colorectal cancer | HB | 350 | 350 | PCR-RFLP |
| Mockelmann, N. | 2009 | German | Colorectal cancer | PB | 1044 | 724 | SNPlex |
| Freire, P. | 2010 | Portuguese | Colorectal cancer | PB | 112 | 152 | Real-time PCR |
| Rigoli, L. | 2010 | Italian | Gastric cancer | PB | 60 | 87 | PCR-RFLP |
| Ashton, K. A. | 2010 | Austrilian | Endometrial cancer | PB | 213 | 291 | Real-time PCR |
| rs2066847/rs5743293 (3020insC) | |||||||
| Nej, K. | 2004 | Polish | Pancreatic cancer | HB | 127 | 300 | Allele-specific PCR |
| Kurzawski, G. | 2004 | Polish | Colorectal cancer | HB | 556 | 300 | Allele-specific PCR |
| Alhopuro, P. | 2004 | Finnish | Colorectal cancer | PB | 926 | 348 | Allele-specific PCR |
| Huzarski, T. | 2005 | Polish | Breast cancer | HB | 462 | 1910 | Allele-specific PCR |
| Debniak, T. | 2005 | Polish | Melanoma | HB | 470 | 649 | Allele-specific PCR |
| Lubinski, J. | 2005 | Polish | Mixed | HB | 2850 | 1910 | Allele-specific PCR |
| Papaconstantinou, I. | 2005 | Greek | Colorectal cancer | N.A. | 104 | 100 | Allele-specific PCR |
| Rothman, N. | 2006 | Caucasian | Non-Hodgkin lymphoma | PB/HB | 3069 | 3497 | Mixed |
| Forrest, M. S. | 2006 | British, American | Non-Hodgkin lymphoma | PB | 899 | 1433 | Taqman |
| Roberts, R. L. | 2006 | New Zealander | Colorectal cancer | PB | 133 | 201 | ARMS |
| Jaworowska, E. | 2006 | Polish | Laryngeal cancer | HB | 347 | 4102 | Allele-specific PCR |
| Irmejs, A. | 2006 | Latvian | Mixed | PB | 420 | 974 | Allele-specific PCR |
| Lener, M. R. | 2006 | Polish | Mixed | HB | 4496 | 2068 | Allele-specific PCR |
| Lakatos, P. L. | 2007 | Hungarian | Colorectal cancer | N.A. | 194 | 200 | dHPLC |
| Vogel, U. | 2007 | Danish | Colorectal cancer | PB | 355 | 753 | CE-SSCP |
| Ennas, M. G. | 2008 | Italian | Lymphocytic leukaemia | PB | 39 | 109 | Taqman |
| Magnowski, P. | 2008 | Polish | Ovarian cancer | HB | 257 | 1910 | Allele-specific PCR |
| Szeliga, J. | 2008 | Polish | Rectal cancer | HB | 51 | 100 | ASA |
| Suchy, J. | 2008 | Polish | Colorectal cancer | HB | 607 | 607 | PCR-RFLP |
| Wex, T. | 2008 | German | Gastric cancer | PB | 47 | 48 | PCR-RFLP |
| Ture-Ozdemir, F. | 2008 | Greek | Gastric MALT lymphoma | HB | 56 | 51 | PCR-RFLP |
| Angeletti, S. | 2009 | Italian | Gastric cancer | PB | 170 | 156 | Multiplex PCR |
| Rigoli, L. | 2010 | Italian | Gastric cancer | PB | 60 | 87 | PCR-RFLP |
| Skibola, C. F. | 2010 | Mixed | Non-Hodgkin lymphoma | PB/HB | 6360 | 6636 | Mixed |
| Freire, P. | 2010 | Portuguese | Colorectal cancer | PB | 112 | 152 | Real-time PCR |
Abbreviations: PB: population-based; HB: hospital-based.
Associations of NOD2 Polymorphisms with Cancer Risks
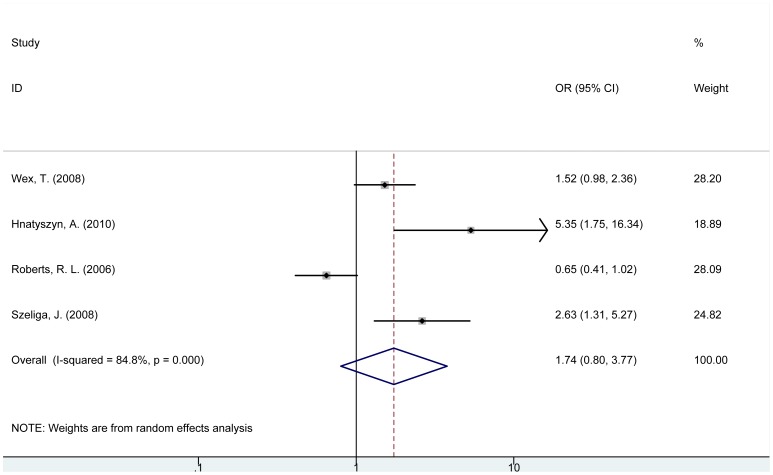
For NOD2 rs2066842 C/T (P268S) polymorphism, carriers of TT or CT genotype were not significantly associated with cancer risk compared with wild-type CC genotype (TT vs. CC: OR = 2.48, 95% CI = 0.85–7.25, P = 0.097; CT vs. CC: OR = 1.32, 95% CI = 0.54–3.25, P = 0.543, Table 2). Similarly, no significant relation was found in recessive effect model of (TT+CT) genotype comparing with CC genotype (OR = 1.74, 95% CI = 0.80–3.77, P = 0.163, Figure 1) or in allele analysis of T allele comparing with C allele (OR = 1.54, 95% CI = 0.74–3.21, P = 0.247) (Table 2). Results from the subgroup analysis of NOD2 rs2066842 polymorphism were presented in Table S1.
Table 2. Meta-analysis results of the association between NOD2 polymorphisms and cancer risks.
| Data set number | OR(95% CI) | P value | Model | Phet | I2(%) | |
| rs2066842 C/T (P268S) | ||||||
| TT vs. CC | 3 | 2.48(0.85–7.25) | 0.097 | R | 0.041 | 68.8% |
| CT vs. CC | 3 | 1.32(0.54–3.25) | 0.543 | R | 0.002 | 83.9% |
| (TT+CT) vs. CC | 4 | 1.74(0.80–3.77) | 0.163 | R | <0.001 | 84.8% |
| T allele vs. C allele | 3 | 1.54(0.74–3.21) | 0.247 | R | <0.001 | 87.6% |
| rs2066844 C/T (R702W) | ||||||
| TT vs. CC | 7 | 3.77(1.30–10.93) | 0.015 | F | 0.970 | 0.0% |
| CT vs. CC | 13 | 1.34(1.01–1.76) | 0.040 | R | 0.033 | 46.5% |
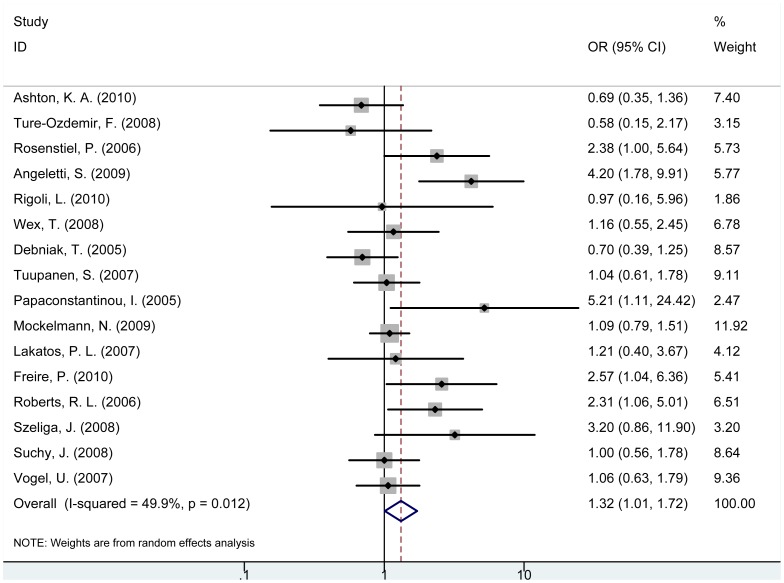
| (TT+CT) vs. CC | 16 | 1.32(1.01–1.72) | 0.041 | R | 0.012 | 49.9% |
| T allele vs. C allele | 13 | 1.43(1.09–1.88) | 0.010 | R | 0.024 | 48.8% |
| rs2066845 C/G (G908R) | ||||||
| CG vs. GG | 11 | 1.39(1.03–1.87) | 0.030 | F | 0.186 | 27.1% |
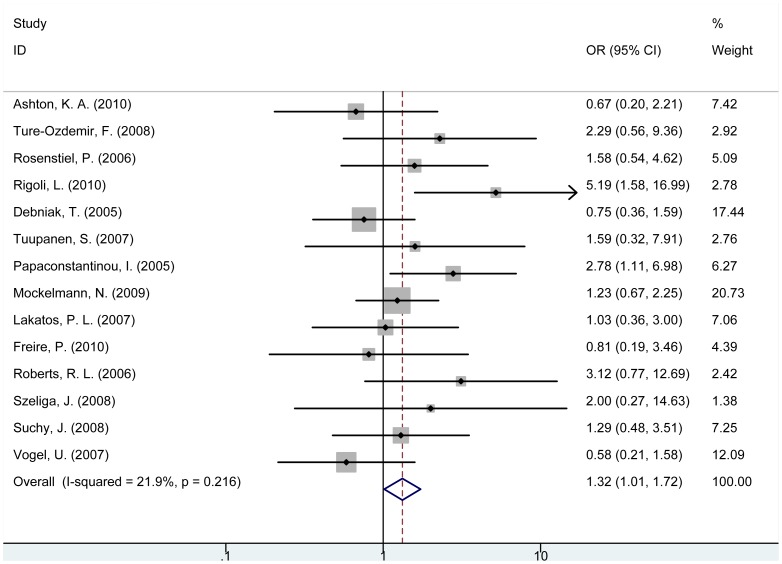
| (CC+CG) vs. GG | 14 | 1.32(1.01–1.72) | 0.040 | F | 0.216 | 21.9% |
| C allele vs. G allele | 11 | 1.40(1.05–1.88) | 0.024 | F | 0.235 | 21.8% |
| rs2066847/rs5743293 (3020insC) | ||||||
| +/+ vs. −/− | 4 | 3.42(1.59–7.40) | 0.002 | F | 0.514 | 0.0% |
| +/− vs. −/− | 14 | 1.35(1.06–1.72) | 0.016 | R | 0.010 | 54.0% |
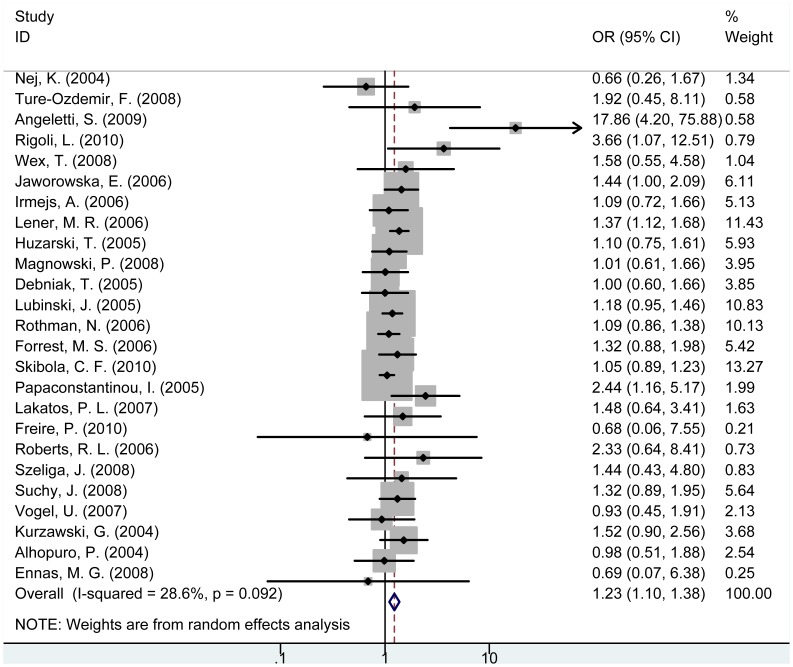
| (+/+ and +/−) vs. −/− | 25 | 1.23(1.10–1.38) | <0.001 | R | 0.092 | 28.6% |
| + vs. − | 13 | 1.40(1.11–1.76) | 0.004 | R | 0.014 | 52.4% |
R: random effect model; F: fixed effect model.
Figure 1. Forest plot for the association between NOD2 rs2066842 polymorphism and cancer risk (TT+CT vs. CC).
For NOD2 rs2066844 C/T (R702W) polymorphism, individuals with TT or CT genotype were associated with increased risk of cancer compared with CC carriers, respectively (TT vs. CC: OR = 3.77, 95% CI = 1.30–10.93, P = 0.015; CT vs. CC: OR = 1.34, 95% CI = 1.01–1.76, P = 0.040, Table 2). (TT+CT) genotype was associated with increased risk of cancer compared with wild-type CC genotype (OR = 1.32, 95% CI = 1.01–1.72, P = 0.041, Figure 2). In the subgroup analysis of cancer type, (TT+CT) genotype was associated with significantly increased risk of CRC (OR = 1.26, 95% CI = 1.03–1.53, P = 0.027) but no significant association was observed for gastric tumors (Table S2). In addition, T allele of NOD2 rs2066844 C/T polymorphism was associated with significantly increased risk of cancer compared with C allele (Table 2).
Figure 2. Forest plot for the association between NOD2 rs2066844 polymorphism and cancer risk (TT+CT vs. CC).
For NOD2 rs2066845 C/G (G908R) polymorphism, CG genotype carriers were observed to be significantly associated with increased risk of cancer compared with GG carriers (OR = 1.39, 95% CI = 1.03–1.87, P = 0.030, Table 2). Individuals with (CC+CG) genotype were significantly associated with increased risk of cancer in the overall analysis (OR = 1.32, 95% CI = 1.01–1.72, P = 0.040, Figure 3) and in gastric tumor subgroup (OR = 2.70, 95% CI = 1.39–5.25, P = 0.003, Table S3), but no significant association was observed in CRC subgroup. Additionally, C allele was associated with increased cancer risk compared with G allele.
Figure 3. Forest plot for the association between NOD2 rs2066845 polymorphism and cancer risk (CC+CG vs. GG).
For NOD2 rs2066847/rs5743293 (3020insC) polymorphism, carriers of insC/insC or insC/− genotype were associated with increased cancer risk compared with wild-type −/− carriers, respectively (insC/insC vs. −/−: OR = 3.42, 95% CI = 1.59–7.40, P = 0.002; insC/− vs. −/−: OR = 1.35, 95% CI = 1.06–1.72, P = 0.016). Individuals with (insC/insC+insC/−) genotype were significantly associated with increased risk of cancer compared with −/− carriers (OR = 1.23, 95% CI = 1.10–1.38, P<0.001, Figure 4). In the subgroup analysis of cancer type, (insC/insC+insC/−) genotype was significantly associated with increased risk of colorectal cancer, gastric cancer and MALT lymphoma, breast cancer, lung cancer, laryngeal cancer but not with urogenital cancer, pancreatic cancer, melanoma or non-Hodgkin lymphoma (Table S4). In the subgroup analysis of control source, (insC/insC+insC/−) genotype was significantly associated with increased cancer risk in hospital-based subgroup (OR = 1.25, 95% CI = 1.12–1.40, P<0.001) but no significant association was observed in population-based subgroup (Table S4).
Figure 4. Forest plot for the association between NOD2 rs2066847 polymorphism and cancer risk ((+/+ and +/−) vs. −/−).
Heterogeneity Test, Sensitivity Analysis, and Publication Bias
For most comparisons for NOD2 gene rs2066844 C/T, rs2066845 C/G and rs2066847 insC polymorphisms, no obvious heterogeneity was observed (I2<50%). The exclusion of each single study did not significantly change the overall outcome, suggesting that the results of the meta-analysis were robust. However, in most comparisons of rs2066842 C/T polymorphism, significant heterogeneity was observed, which could not be fully explained by study design or subgroup analysis. The heterogeneity might result from the limited number of studies included. Besides, meta-regression was not performed to explore the source of the heterogeneity due to the limited study number.
The Begg’s test and Egger’s test were performed to quantitatively evaluate the publication bias of the studies. The detailed results for publication bias test were summarized in Table 3. No significant publication bias was observed in this meta-analysis except two comparisons in NOD2 rs2066847 insC polymorphism. In addition, funnel plots that qualitatively evaluated the publication bias of association between NOD2 rs2066847 insC polymorphism and cancer risk was presented in Figure S2.
Table 3. Publication bias.
| Compared genotype | Begg's test | Egger's test | |||
| z value | P value | t value | P value | ||
| rs2066842 C/T (P268S) | TT vs. CC | 0.52 | 0.602 | 1.35 | 0.405 |
| CT vs. CC | 0.52 | 0.602 | 0.87 | 0.546 | |
| (TT+CT) vs. CC | 1.36 | 0.174 | 1.70 | 0.231 | |
| T allele vs. C allele | 0.52 | 0.602 | 1.04 | 0.488 | |
| rs2066844 C/T (R702W) | TT vs. CC | -0.75 | 0.453 | 0.00 | 0.997 |
| CT vs. CC | 1.83 | 0.067 | 1.96 | 0.076 | |
| (TT+CT) vs. CC | 1.53 | 0.126 | 1.79 | 0.095 | |
| T allele vs. C allele | 1.95 | 0.051 | 2.00 | 0.071 | |
| rs2066845 C/G (G908R) | CG vs. GG | 0.39 | 0.697 | 0.45 | 0.666 |
| (CC+CG) vs. GG | 0.93 | 0.352 | 1.05 | 0.313 | |
| C allele vs. G allele | 0.08 | 0.938 | 0.26 | 0.800 | |
| rs2066847/rs5743293 (3020insC) | +/+ vs. −/− | 0.68 | 0.497 | 1.28 | 0.329 |
| +/− vs. −/− | 1.46 | 0.143 | 2.53 | 0.028 | |
| (+/+ and +/−) vs. −/− | 1.03 | 0.304 | 1.88 | 0.073 | |
| + vs. − | 1.10 | 0.272 | 2.42 | 0.034 | |
Discussion
Results from previous individual studies investigating the associations between NOD2 polymorphisms and cancer risk were inconclusive. To our knowledge, this was the first comprehensive meta-analysis concerning the effect of NOD2 rs2066842 C/T, rs2066844 C/T, rs2066845 C/G and rs2066847 insC polymorphisms on risks of overall cancer and specific cancer subtypes. By analyzing the data extracted from 30 full-text publications, we revealed that NOD2 rs2066844 C/T, rs2066845 C/G and rs2066847 (3020insC) polymorphisms might be associated with increased cancer risk especially for gastrointestinal cancer but no significant association was observed between NOD2 rs2066842 C/T polymorphism and cancer risk.
NOD2 gene comprises 12 exons and encodes a protein consisting of 1040 amino acids. NOD2 recognized microbial pathogens located in the cytoplasm through the specific detection of conserved muramyl dipeptides and induced nuclear factor kappa B (NF-κB) activation via the RIP2/IKK pathway [43]. The NF-κB pathway acts to enhance the expression of proinflammatory molecules, thereby stimulating both adaptive and innate immune responses. In addition, NOD2 was implicated in programmed cell death and was known to be key regulator of chronic inflammatory conditions [44]. Recent attention has been given to the role of NOD2 polymorphisms in carcinogenesis. Of which, four polymorphisms (rs2066842 C/T, rs2066844 C/T, rs2066845 C/G and rs2066847 insC) were of great interest. However, results of the individual studies came up with inconsistent conclusions.
In this meta-analysis, NOD2 rs2066842 polymorphism was not observed to be associated with cancer risk in all comparisons. Only four studies were analyzed in the pooled estimates and obvious heterogeneities were detected which could not be explained by subgroup analysis or solved by meta-regression. Therefore, further large-scale studies were required to validate the results. With respect to NOD2 rs2066844, rs2066845 and rs2066847 polymorphisms, the dominant effect models of the three polymorphisms all indicated significantly increased cancer risk (OR for rs2066844 = 1.32; OR for rs2066845 = 1.32; OR for rs2066847 = 1.23) (Table 2). The allele analysis found consistently increased cancer risk (OR for rs2066844 = 1.43; OR for rs2066845 = 1.40; OR for rs2066847 = 1.40) (Table 2).
Different cancer has its distinct mechanisms of initiation and progression, in which polymorphisms of key genes play a critical role. The present meta-analysis unraveled that NOD2 polymorphisms were observed of different associations with cancer in various cancer subgroups. The (TT+CT) genotype of rs2066844 conferred increased risk to CRC but not gastric cancer and MALT lymphoma; individuals with (CC+CG) genotype of rs2066845 were associated with higher risk of gastric cancer and MALT lymphoma rather than CRC; (insC/insC+insC/−) genotype carriers of rs2066847 were associated with increased risk of colorectal cancer, gastric cancer and MALT lymphoma, breast cancer, lung cancer, laryngeal cancer but not with urogenital cancer, pancreatic cancer, melanoma or non-Hodgkin lymphoma. Besides, the source of controls adopted would influence the associations of polymorphisms with cancer risks. In the present study, (insC/insC+insC/−) genotype carriers of rs2066847 were associated with increased cancer risk in subgroup of hospital-based but not in population-based. The above-mentioned results of subgroup analysis uncovered underlying information and deserved future concerns.
Inflammation may lead to carcinogenesis by stimulating continuous cell proliferation, inducing DNA damage, and arousing angiogenesis [45]. Mutations of NOD2 gene have been linked to a number of chronic inflammatory diseases including Crohn’s disease, atopic dermatitis and so on [46]. As NOD2 is implicated in microbial recognition and inflammatory reactions, the association of NOD2 polymorphisms with cancer risk might due to the alteration of the ability of inducing immune response to bacteria which consequently results in the development of persistent bacterial infection or enhanced production of proinflammatory mediators. The three polymorphisms (rs2066844, rs2066845 and rs2066847) which were observed to be associated with increased risk of cancer in this meta-analysis were all located at the leucine-rich region (LRR) of NOD2 protein. The amino acid substitutions caused by these polymorphisms would alter protein function or splicing, thus influencing the role of NOD2 in the regulation of apoptosis and chronic inflammation and finally leading to cancer. As for functional studies, 3020insC variant, leading to a substitution in the 10th LRR followed by a premature stop codon, has been proved to be less active in the response to bacterial lipopolysaccharides, which might produce an increased inflammatory response [47]. Mice deficient in NOD1, NOD2, or RIPK2 exhibited increased susceptibility to bacteria, which arises from a decreased ability to recruit neutrophils and less production in proinflammatory and antimicrobial molecules [17], [48]. Although the above-mentioned studies could, at least in part, explain the observed relation of NOD2 polymorphisms with cancer risk, future investigations concerning the specific mechanism of NOD2 polymorphisms in carcinogenesis are still required.
Several limitations should be acknowledged in this meta-analysis when interpreting the results. First, the sample size was not sufficiently large for the pooled analysis of NOD2 rs2066842 C/T polymorphism and some subgroup analyses for NOD2 rs2066844 C/T, rs2066845 C/G and rs2066847 insC polymorphisms. Second, all the included studies in the current meta-analysis were published in English, therefore publication bias might exist although the statistical test did not indicate it. Third, the ethnicities of all the available studies were Caucasian populations, which inevitably limited the generalizability of our conclusion on other populations. Fourth, obvious heterogeneities were observed in a few comparisons, which would limit the accuracy of certain associations. Finally, as other important data of environment factors such as smoking or drinking were not available for individual studies, we could not obtained results with adjustments by other co-variables.
Conclusion
To be concluded, this meta-analysis suggested that NOD2 rs2066844 C/T, rs2066845 C/G and rs2066847 (3020insC) polymorphisms might be associated with increased cancer risk especially for gastrointestinal cancer. No significant association was observed between NOD2 rs2066842 C/T polymorphism and cancer risk. Further large-scale and well-designed studies concerning different ethnicities are still needed to confirm the results of our meta-analysis.
Supporting Information
The flowchart of literature inclusion and exclusion.
(TIF)
Funnel plot for studies of association between NOD2 rs2066847 polymorphism and cancer risk ((+/+ and +/−) vs. −/−).
(TIF)
Subgroup analysis of association between NOD2 rs2066842 polymorphism and cancer risk.
(DOC)
Subgroup analysis of association between NOD2 rs2066844 polymorphism and cancer risk.
(DOC)
Subgroup analysis of association between NOD2 rs2066845 polymorphism and cancer risk.
(DOC)
Subgroup analysis of association between NOD2 rs2066847 polymorphism and cancer risk.
(DOC)
PRISMA checklist.
(DOC)
Funding Statement
This study is supported by grants from National Basic Research Program of China (973 Program Ref No.2010CB529304), the grants of the Science and Technology Project of Liaoning province (Ref No.2011225002) and the grants of the Science and Technology Project of Liaoning province (Ref No.2012225016). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Thun MJ, DeLancey JO (2010) Center MM, Jemal A, Ward EM (2010) The global burden of cancer: priorities for prevention. Carcinogenesis 31: 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zaridze DG (2008) Molecular epidemiology of cancer. Biochemistry (Mosc) 73: 532–542. [DOI] [PubMed] [Google Scholar]
- 4. Deng W, Xie J (2012) NOD2 signaling and role in pathogenic mycobacterium recognition, infection and immunity. Cell Physiol Biochem 30: 953–963. [DOI] [PubMed] [Google Scholar]
- 5. Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, et al. (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem 278: 8869–8872. [DOI] [PubMed] [Google Scholar]
- 6. Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, et al. (2001) Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411: 599–603. [DOI] [PubMed] [Google Scholar]
- 7. Gazouli M, Mantzaris G, Kotsinas A, Zacharatos P, Papalambros E, et al. (2005) Association between polymorphisms in the Toll-like receptor 4, CD14, and CARD15/NOD2 and inflammatory bowel disease in the Greek population. World J Gastroenterol 11: 681–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kurzawski G, Suchy J, Kladny J, Grabowska E, Mierzejewski M, et al. (2004) The NOD2 3020insC mutation and the risk of colorectal cancer. Cancer Res 64: 1604–1606. [DOI] [PubMed] [Google Scholar]
- 9. Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 10. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 11. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 12. Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50: 1088–1101. [PubMed] [Google Scholar]
- 13. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. Bmj 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ashton KA, Proietto A, Otton G, Symonds I, McEvoy M, et al. (2010) Toll-like receptor (TLR) and nucleosome-binding oligomerization domain (NOD) gene polymorphisms and endometrial cancer risk. BMC Cancer 10: 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nej K, Bartsch DK, Sina-Frey M, Rieder H, Hahn SA, et al. (2004) The NOD2 3020insC Mutation and The Risk of Familial Pancreatic Cancer? Hered Cancer Clin Pract 2: 149–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ture-Ozdemir F, Gazouli M, Tzivras M, Panagos C, Bovaretos N, et al. (2008) Association of polymorphisms of NOD2, TLR4 and CD14 genes with susceptibility to gastric mucosa-associated lymphoid tissue lymphoma. Anticancer Res 28: 3697–3700. [PubMed] [Google Scholar]
- 17. Rosenstiel P, Hellmig S, Hampe J, Ott S, Till A, et al. (2006) Influence of polymorphisms in the NOD1/CARD4 and NOD2/CARD15 genes on the clinical outcome of Helicobacter pylori infection. Cell Microbiol 8: 1188–1198. [DOI] [PubMed] [Google Scholar]
- 18. Angeletti S, Galluzzo S, Santini D, Ruzzo A, Vincenzi B, et al. (2009) NOD2/CARD15 polymorphisms impair innate immunity and increase susceptibility to gastric cancer in an Italian population. Hum Immunol 70: 729–732. [DOI] [PubMed] [Google Scholar]
- 19. Rigoli L, Di Bella C, Fedele F, Procopio V, Amorini M, et al. (2010) TLR4 and NOD2/CARD15 genetic polymorphisms and their possible role in gastric carcinogenesis. Anticancer Res 30: 513–517. [PubMed] [Google Scholar]
- 20. Wex T, Ebert MP, Kropf S, Dierkes J, Schuttler K, et al. (2008) Gene polymorphisms of the NOD-2/CARD-15 gene and the risk of gastric cancer in Germany. Anticancer Res 28: 757–762. [PubMed] [Google Scholar]
- 21. Hnatyszyn A, Szalata M, Stanczyk J, Cichy W, Slomski R (2010) Association of c.802C>T polymorphism of NOD2/CARD15 gene with the chronic gastritis and predisposition to cancer in H. pylori infected patients. Exp Mol Pathol 88: 388–393. [DOI] [PubMed] [Google Scholar]
- 22. Jaworowska E, Masojc B, Tarnowska C, Brzosko M, Flicinski J, et al. (2006) Association between early-onset breast and laryngeal cancers. Breast Cancer Res Treat 97: 215–219. [DOI] [PubMed] [Google Scholar]
- 23. Irmejs A, Miklasevics E, Boroschenko V, Gardovskis A, Vanags A, et al. (2006) Pilot study on low penetrance breast and colorectal cancer predisposition markers in latvia. Hered Cancer Clin Pract 4: 48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lener MR, Oszutowska D, Castaneda J, Kurzawski G, Suchy J, et al. (2006) Prevalence of the NOD2 3020insC mutation in aggregations of breast and lung cancer. Breast Cancer Res Treat 95: 141–145. [DOI] [PubMed] [Google Scholar]
- 25. Huzarski T, Lener M, Domagala W, Gronwald J, Byrski T, et al. (2005) The 3020insC allele of NOD2 predisposes to early-onset breast cancer. Breast Cancer Res Treat 89: 91–93. [DOI] [PubMed] [Google Scholar]
- 26. Magnowski P, Medrek K, Magnowska M, Stawicka M, Kedzia H, et al. (2008) The 3020insC NOD2 gene mutation in patients with ovarian cancer. Ginekol Pol 79: 544–549. [PubMed] [Google Scholar]
- 27. Debniak T, Kurzawski G, Huzarski T, Byrski T, Gronwald J, et al. (2005) NOD2 variants and the risk of malignant melanoma. Eur J Cancer Prev 14: 143–146. [DOI] [PubMed] [Google Scholar]
- 28. Lubinski J, Huzarski T, Kurzawski G, Suchy J, Masojc B, et al. (2005) The 3020insC Allele of NOD2 Predisposes to Cancers of Multiple Organs. Hered Cancer Clin Pract 3: 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Skibola CF, Bracci PM, Nieters A, Brooks-Wilson A, de Sanjose S, et al. (2010) Tumor necrosis factor (TNF) and lymphotoxin-alpha (LTA) polymorphisms and risk of non-Hodgkin lymphoma in the InterLymph Consortium. Am J Epidemiol 171: 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rothman N, Skibola CF, Wang SS, Morgan G, Lan Q, et al. (2006) Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph Consortium. Lancet Oncol 7: 27–38. [DOI] [PubMed] [Google Scholar]
- 31. Forrest MS, Skibola CF, Lightfoot TJ, Bracci PM, Willett EV, et al. (2006) Polymorphisms in innate immunity genes and risk of non-Hodgkin lymphoma. Br J Haematol 134: 180–183. [DOI] [PubMed] [Google Scholar]
- 32. Papaconstantinou I, Theodoropoulos G, Gazouli M, Panoussopoulos D, Mantzaris GJ, et al. (2005) Association between mutations in the CARD15/NOD2 gene and colorectal cancer in a Greek population. Int J Cancer 114: 433–435. [DOI] [PubMed] [Google Scholar]
- 33. Mockelmann N, von Schonfels W, Buch S, von Kampen O, Sipos B, et al. (2009) Investigation of innate immunity genes CARD4, CARD8 and CARD15 as germline susceptibility factors for colorectal cancer. BMC Gastroenterol 9: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lakatos PL, Hitre E, Szalay F, Zinober K, Fuszek P, et al. (2007) Common NOD2/CARD15 variants are not associated with susceptibility or the clinicopathologic characteristics of sporadic colorectal cancer in Hungarian patients. BMC Cancer 7: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Freire P, Portela F, Donato MM, Figueiredo P, Ferreira M, et al. (2010) CARD15 mutations and colorectal cancer in a South European country. Int J Colorectal Dis 25: 1211–1219. [DOI] [PubMed] [Google Scholar]
- 36. Roberts RL, Gearry RB, Allington MD, Morrin HR, Robinson BA, et al. (2006) Caspase recruitment domain-containing protein 15 mutations in patients with colorectal cancer. Cancer Res 66: 2532–2535. [DOI] [PubMed] [Google Scholar]
- 37. Tuupanen S, Alhopuro P, Mecklin JP, Jarvinen H, Aaltonen LA (2007) No evidence for association of NOD2 R702W and G908R with colorectal cancer. Int J Cancer 121: 76–79. [DOI] [PubMed] [Google Scholar]
- 38. Szeliga J, Sondka Z, Jackowski M, Jarkiewicz-Tretyn J, Tretyn A, et al. (2008) NOD2/CARD15 polymorphism in patients with rectal cancer. Med Sci Monit 14: CR480–484. [PubMed] [Google Scholar]
- 39. Suchy J, Klujszo-Grabowska E, Kladny J, Cybulski C, Wokolorczyk D, et al. (2008) Inflammatory response gene polymorphisms and their relationship with colorectal cancer risk. BMC Cancer 8: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vogel U, Christensen J, Dybdahl M, Friis S, Hansen RD, et al. (2007) Prospective study of interaction between alcohol, NSAID use and polymorphisms in genes involved in the inflammatory response in relation to risk of colorectal cancer. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis 624: 88–100. [DOI] [PubMed] [Google Scholar]
- 41. Alhopuro P, Ahvenainen T, Mecklin JP, Juhola M, Jarvinen HJ, et al. (2004) NOD2 3020insC alone is not sufficient for colorectal cancer predisposition. Cancer Res 64: 7245–7247. [DOI] [PubMed] [Google Scholar]
- 42. Ennas MG, Moore PS, Zucca M, Angelucci E, Cabras MG, et al. (2008) Interleukin-1B (IL1B) and interleukin-6 (IL6) gene polymorphisms are associated with risk of chronic lymphocytic leukaemia. Hematol Oncol 26: 98–103. [DOI] [PubMed] [Google Scholar]
- 43. Kersse K, Bertrand MJ, Lamkanfi M, Vandenabeele P (2011) NOD-like receptors and the innate immune system: coping with danger, damage and death. Cytokine Growth Factor Rev 22: 257–276. [DOI] [PubMed] [Google Scholar]
- 44. Strober W, Watanabe T (2011) NOD2, an intracellular innate immune sensor involved in host defense and Crohn's disease. Mucosal Immunol 4: 484–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jackson AL, Loeb LA (2001) The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res 477: 7–21. [DOI] [PubMed] [Google Scholar]
- 46. Zhong Y, Kinio A, Saleh M (2013) Functions of NOD-Like Receptors in Human Diseases. Front Immunol 4: 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, et al. (2001) A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411: 603–606. [DOI] [PubMed] [Google Scholar]
- 48. Frutuoso MS, Hori JI, Pereira MS, Junior DS, Sonego F, et al. (2010) The pattern recognition receptors Nod1 and Nod2 account for neutrophil recruitment to the lungs of mice infected with Legionella pneumophila. Microbes Infect 12: 819–827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The flowchart of literature inclusion and exclusion.
(TIF)
Funnel plot for studies of association between NOD2 rs2066847 polymorphism and cancer risk ((+/+ and +/−) vs. −/−).
(TIF)
Subgroup analysis of association between NOD2 rs2066842 polymorphism and cancer risk.
(DOC)
Subgroup analysis of association between NOD2 rs2066844 polymorphism and cancer risk.
(DOC)
Subgroup analysis of association between NOD2 rs2066845 polymorphism and cancer risk.
(DOC)
Subgroup analysis of association between NOD2 rs2066847 polymorphism and cancer risk.
(DOC)
PRISMA checklist.
(DOC)