Summary
Growth of prostate cancer cells is dependent upon androgen stimulation of the androgen receptor (AR). Dihydrotestosterone (DHT), the most potent androgen, is usually synthesized in the prostate from testosterone secreted by the testis. Following chemical or surgical castration, prostate cancers usually shrink owing to testosterone deprivation. However, tumors often recur, forming castration-resistant prostate cancer (CRPC). Here, we show that CRPC sometimes expresses a gain-of-stability mutation leading to a gain-of-function in 3β-hydroxysteroid dehydrogenase type 1 (3βHSD1), which catalyzes the initial rate-limiting step in the conversion of the adrenal-derived steroid dehydroepiandrosterone to DHT. The mutation (N367T) does not affect catalytic function, but it renders the enzyme resistant to ubiquitination and degradation, leading to profound accumulation. Whereas dehydroepiandrosterone conversion to DHT is usually very limited, expression of 367T accelerates this conversion and provides the DHT necessary to activate the AR. We suggest that 3βHSD1 is a valid target for the treatment of CRPC.
Introduction
The growth of cancerous prostate cells requires stimulation of the androgen receptor (AR) by androgens, the most potent of which is dihydrotestosterone (DHT). Advanced prostate cancer usually initially regresses with gonadal testosterone (T) deprivation therapy (i.e., medical or surgical castration), but it almost always eventually progresses as castration-resistant prostate cancer (CRPC) (Attard et al., 2009; Penning, 2010; Scher and Sawyers, 2005; Sharifi et al., 2005; Yuan and Balk, 2009). The CRPC phenotype is driven by a gain-of-function in the androgen receptor (AR) that is usually accompanied by intratumoral DHT concentrations of about 1 nM, which is sufficient to drive expression of AR-induced genes, including the TMPRSS2-ETS fusion oncogene (Geller et al., 1978; Luu-The et al., 2008; Montgomery et al., 2008; Sharifi, 2013; Titus et al., 2005; Tomlins et al., 2005). The requirement for intratumoral androgen synthesis in driving CRPC progression is most clearly demonstrated by the survival benefit conferred by abiraterone acetate, a drug which blocks androgen synthesis by inhibiting 17α-hydroxylase/17,20-lyase (CYP17A1), and enzalutamide, a potent AR antagonist that blocks DHT access to the AR ligand-binding domain (Barrie et al., 1994; de Bono et al., 2011; Scher et al., 2012; Tran et al., 2009). Intratumoral synthesis of DHT from precursors that are secreted from the adrenal gland occurs through a pathway that circumvents T (Chang et al., 2011). This synthesis requires three enzymes: 3β-hydroxysteroid dehydrogenase (3βHSD; encoded by HSD3B), steroid-5α-reductase (SRD5A) and 17β-hydroxysteroid dehydrogenase (17βHSD) isoenzymes (see Fig 1A) (Chang et al., 2011; Knudsen and Penning, 2010). Nonetheless, increased DHT synthesis in CRPC has not yet been ascribed to any mutations in genes encoding components of the steroidogenic machinery. 3βHSD oxidizes 3β-hydroxyl to 3-keto and isomerizes Δ5 to Δ4 (see Fig 1A), reactions that together make this step practically irreversible by an enzyme that is required for all possible pathways that lead to the synthesis of DHT (Evaul et al., 2010). HSD3B1 encodes for the peripherally expressed isoenzyme (3βHSD1) and has a germline single nucleotide polymorphism (SNP) at position 1245 of HSD3B1, converting A → C, which exchanges an asparagine (N) for a threonine (T) at 3βHSD1 amino acid position 367.
Fig. 1.
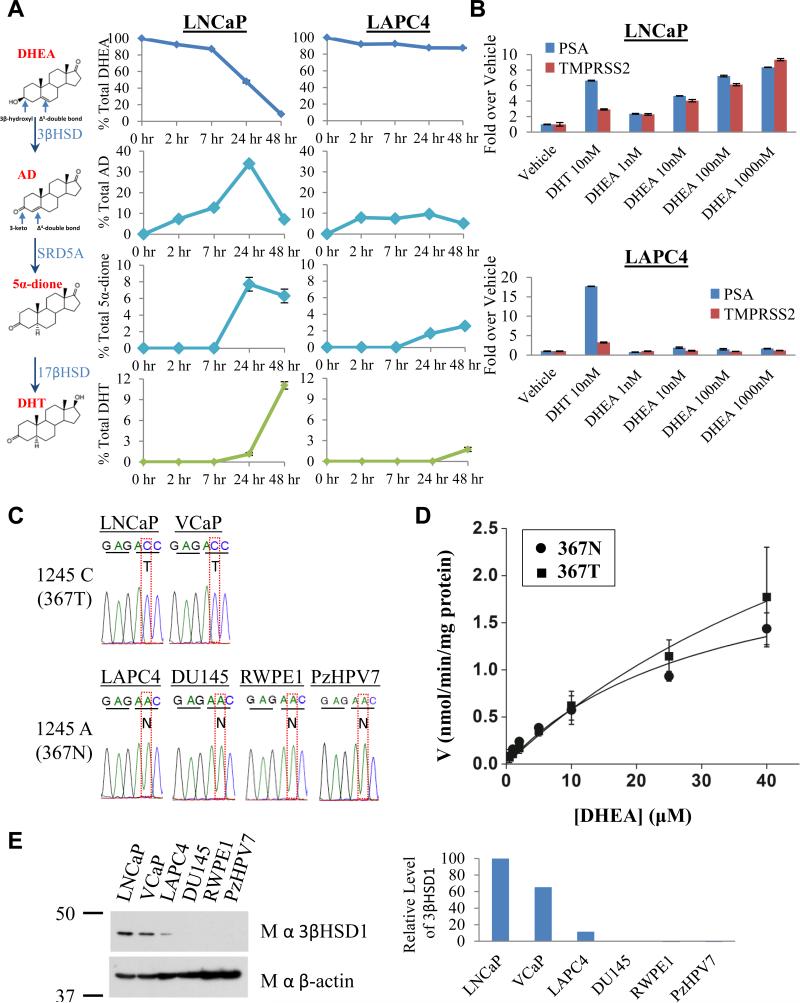
The 3βHSD1(367T) protein encoded by mutant HSD3B1(1245C) increases flux from DHEA to AD, which is otherwise rate-limiting, en route to DHT and expression of AR-responsive genes. (A) Metabolic flux from [3H]-DHEA (100 nM) to AD and downstream to 5α-dione and DHT is robust in LNCaP but limited in LAPC4. The metabolic pathway and steroid structures are shown, indicating sites of modification by 3βHSD1 in converting DHEA to AD. Steroids were quantitated at the indicated time points by HPLC. (B) DHEA induces PSA and TMPRSS2 expression in a concentration-dependent manner in LNCaP but not LAPC4. Expression was assessed by qPCR and normalized to RPLP0 and vehicle control. (C) A substitution converting A → C at position 1245 in HSD3B1 occurs in LNCaP and VCaP encoding a change from N → T at amino acid 367 in 3βHSD1. (D) Wild-type 3βHSD1(367N) and 3βHSD1(367T) have comparable kinetic properties. Michaelis-Menten plot of DHEA metabolism with 3βHSD1(367N) (circle) and 3βHSD1(367T) (square) enzyme. The Km for 3βHSD1(367N) and 3βHSD1(367T) protein are 32 and 77 μM, respectively. (E) Endogenous expression of 3βHSD1(367T) is associated with increased protein quantity. Error bars in A, B and D represent the SD from experiments performed in triplicate. See also Figure S1.
Here, we show that CRPC sometimes expresses the 367T form of 3βHSD1 (3βHSD1(367T)), which increases metabolic flux from dehydroepiandrosterone (DHEA) via the 5α-androstanedione (5α-dione) pathway to DHT by protein resistance to ubiquitination and degradation rather than increased catalytic activity. Selection for 3βHSD1(367T) is evident from somatic mutation in human CRPC tumors, by loss-of-heterozygosity (LOH) of the wild-type copy in patients with germline heterozygous inheritance and from the generation and expression of the same somatic mutation occurring in a mouse xenograft model treated with abiraterone acetate.
Results
Cells with 3βHSD1(367T) have increased flux to DHT
Conversion of DHEA by 3βHSD1 to Δ4-androstenedione (AD) is a proximal step in peripheral tissues for metabolism from adrenal precursors to DHT (Lorence et al., 1990; Simard et al., 2005). Two cell lines derived from patients with CRPC have widely disparate flux from DHEA to AD (Fig 1A), despite comparable expression of transcripts encoding both 3βHSD1 and 3βHSD2 (Fig S1A). Under the same conditions, LNCaP cells metabolize > 90% of [3H]-DHEA by 3βHSD enzymatic activity to AD after 48 hours, whereas LAPC4 cells metabolize only approximately 10% of [3H]-DHEA. In LAPC4 but not LNCaP, apparent rate-limiting conversion of DHEA to AD en route to DHT via the dominant pathway (DHEA → AD → 5α-dione → DHT) (Chang et al., 2011) is further evident by limited accumulation of downstream metabolites and absence of DHEA concentration-dependent increases in AR-regulated PSA and TMPRSS2 (Fig 1B). Sequencing the exons of both HSD3B isoenzymes reveals a single nonsynonymous substitution (Fig 1C) at position 1245 of HSD3B1, converting A → C and exchanges an asparagine (N) for a threonine (T) at 3βHSD1 amino acid position 367 in LNCaP but not LAPC4. To further test the association between HSD3B1 sequence and steroid metabolism, other human prostate cell lines were investigated. Presence of wild-type (1245A) and variant (1245C) HSD3B1 sequence in other prostate cancer and immortalized prostate cell lines is also concordant with “slow” and “fast” flux from DHEA to AD, respectively (Fig S1B). The kinetic properties of recombinant 3βHSD1(367N) and 3βHSD1(367T) proteins, however, do not explain the differences in steroid metabolism between cells expressing each protein (Fig 1D). Western blot was performed to determine if the allele encoding 3βHSD1(367T) is associated with a greater amount of protein in these cells. Both models that encode for 3βHSD1(367T) have increased 3βHSD1 protein compared with the models that have wild-type sequence (Fig 1E).
Androgen deprivation selects for HSD3B1 (1245C)
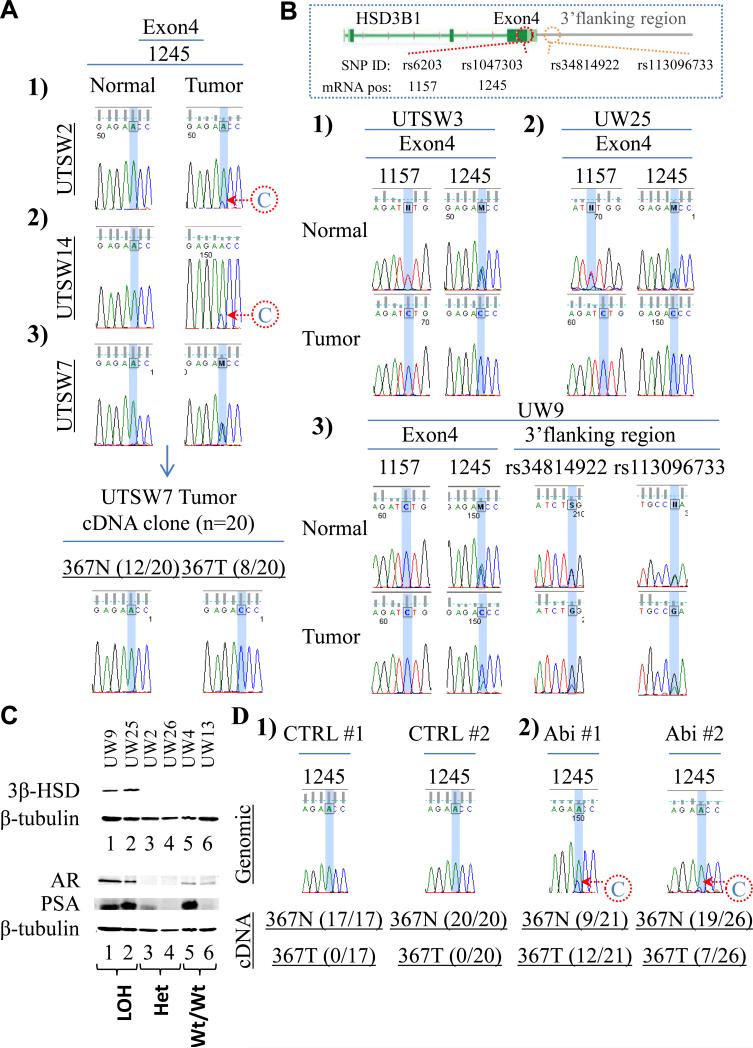
The HSD3B1(1245C); 3βHSD1(367T) allele occurs as a germline SNP variant (rs1047303; 22% allele frequency) (Shimodaira et al., 2010) but might also occur as a somatic mutation in prostate cancer. Although germline homozygous HSD3B1(1245C) inheritance cannot be ruled out, the most likely scenarios accounting for the sole presence of the HSD3B1(1245C) allele evident in both LNCaP and VCaP, given the low expected frequency of homozygous HSD3B1(1245C) inheritance, are either germline heterozygous inheritance followed by loss-of-heterozygosity (LOH) of the wild-type allele, or germline homozygous wild-type inheritance followed by somatic mutation of 1245 A → C. To identify the existence of these possible mechanisms of HSD3B1(1245C) selection in human tumors, matching germline and tumor DNA were sequenced from men with CRPC. Genomic DNA was isolated from CRPC and normal tissue from patients treated at the University of Texas Southwestern Medical Center (UTSW) and from the University of Washington (UW) rapid autopsy program (Montgomery et al., 2008). Patient and tumor characteristics are available in Table S1. Of 40 men with CRPC, the germline of 25, 11 and 4 individuals are homozygous wild-type HSD3B1(1245A), heterozygous and homozygous variant HSD3B1(1245C), respectively. Three of 25 (12%) CRPC tumors with homozygous HSD3B1(1245A) inheritance have acquired the HSD3B1(1245C) allele (Fig 2A). Expression of HSD3B1(1245C) transcript was confirmed in the one available fresh-frozen tumor. It is highly likely that the observation of 3 identical de novo mutations occurring in 25 patients is due to selection rather than chance alone, with a high degree of statistical significance (p=1.47 × 10−13), using the binomial method and assuming a mutation rate of 4 per 1,000,000 base pairs (Greenman et al., 2007). Of 11 CRPC tumors with heterozygous inheritance, 3 (27%) have LOH of the HSD3B1(1245A) allele, resulting in the HSD3B1(1245C) allele being predominantly detectable (Fig 2B). In these three tumors, LOH of adjacent heterozygous SNPs further confirms loss of this region of chromosome 1. In contrast, none of the 11 cases with heterozygous inheritance exhibited LOH of the HSD3B1(1245C) allele (Fig S2A).
Fig. 2.
Somatic selection for HSD3B1(1245C) encoding 3βHSD1(367T) occurs with resistance to androgen deprivation. (A) Conversion from A → C in HSD3B1 occurs in 3 CRPC tumors from patients with homozygous wild-type inheritance. Sequence of cDNA clones from a fresh-frozen tumor (UTSW7) confirms expression of HSD3B1(1245C) transcript. (B) Three CRPC tumors from patients with heterozygous inheritance exhibit LOH of the wild-type HSD3B1(1245A) allele. Sequencing informative (heterozygous) adjacent 5’ (rs6203) and 3’ (rs34814922 and rs113096733) SNPs confirms LOH. (C) 3βHSD1 protein is abundant in tumors with LOH of the HSD3B1(1245A) allele but not tumors with heterozygous expression or homozygous HSD3B1(1245A) expression. Both tumors with LOH tested also express AR and PSA (20 μg protein loaded per lane for each tumor). (D) Somatic mutation converting A → C in HSD3B1 occurs in two LAPC4 xenograft tumors treated with abiraterone acetate (Abi) after orchiectomy and expression of HSD3B1(1245C) transcript encoding 3βHSD1(367T) is evidenced by sequencing cDNA clones from these tumors. Genomic sequence from two representative control tumors (CTRL#1 and CTRL#2) treated with orchiectomy alone is shown for comparison. All 37 cDNA clones from CTRL#1 and CTRL#2 have HSD3B1(1245A) transcript encoding 3βHSD1(367N). See also Figure S2 and Table S1.
Two tumors (UW9 and UW25) with LOH of the HSD3B1(1245A) allele had tissue remaining for additional studies. Consistent with the findings in LNCaP and VCaP that only have the HSD3B1(1245C) allele, both of these tumors have abundant detectable 3βHSD1 protein (Fig 2C). In contrast, both tumors tested with heterozygous expression and homozygous HSD3B1(1245A) expression have little or no detectable 3βHSD1. mRNA quantitation by qPCR demonstrates that the increased 3βHSD1 protein abundance occurring specifically in the tumors with LOH is not attributable to transcript overexpression (Fig S2B). Both tumors with LOH robustly express AR and PSA, suggesting that flux to DHT sustained by 3βHSD1 protein functions to elicit AR signaling (Fig 2C).
Abiraterone inhibits CYP17A1 and weakly inhibits 3βHSD, further decreasing intratumoral androgen concentrations and extending survival in CRPC (de Bono et al., 2011; Li et al., 2012); therefore, conversion to the HSD3B1(1245C) allele encoding 3βHSD1(367T) might permit sustained androgen synthesis despite lower availability of precursors. To determine if abiraterone treatment selects for the HSD3B1(1245C) allele, genomic DNA from LAPC4 xenograft tumors grown in orchiectomized mice treated with abiraterone or vehicle (n=8 mice per treatment) was isolated and sequenced (Li et al., 2012). The 1245C allele is detectable in 2 of 8 tumors (Abi #1 and Abi #2) in the abiraterone treatment group and no tumors in the vehicle group (Fig 2D). To confirm expression of the somatically acquired mutation in the abiraterone group, cDNA clones were generated and sequenced. The mutant HSD3B1(1245C) transcript encoding for 3βHSD1(367T) is confirmed in 12 of 21(57%) cDNA clones sequenced from Abi #1 and 7 of 26 (27%) clones from Abi #2. In contrast, the mutant transcript is not present in any of the 37 cDNA clones obtained from two vehicle treated LAPC4 xenograft tumors.
Blocking 3βHSD1(367T) inhibits DHT synthesis, the AR-response and CRPC
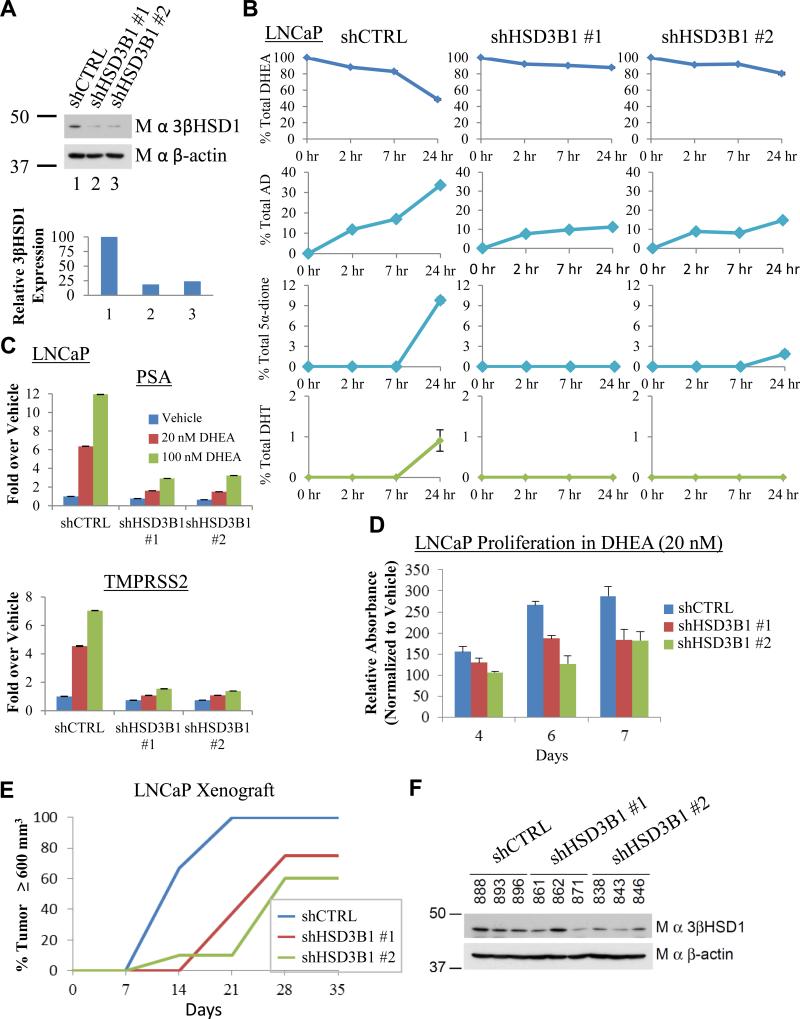
To determine the role of 3βHSD1(367T) expression in regulating flux from DHEA to DHT and AR stimulation, endogenous expression was silenced in LNCaP using 2 independent lentiviral shRNAs (Fig 3A). Blocking 3βHSD1 expression with both shRNAs inhibits flux from DHEA to AD, resulting in little or no detectable conversion to downstream 5α-dione and DHT (Fig 3B). Silencing mutant 3βHSD1 expression and blocking flux to DHT impedes the expression of AR-regulated PSA and TMPRSS2 (Fig 3C), leading to inhibition of cell proliferation in vitro (Fig 3D). In vivo, depletion of endogenously expressed mutant 3βHSD1 significantly hinders CRPC growth in surgically orchiectomized mice (Fig 3E). CRPC tumors that eventually develop from cell lines initially expressing lentiviral shRNA knockdown constructs regain 3βHSD1 protein, probably from selection for cells that have lost the shRNA construct (Fig 3F).
Fig. 3.
Genetic silencing of 3βHSD1(367T) impedes conversion of DHEA to DHT, induction of PSA and TMPRSS2 expression, and CRPC growth. (A) Stable lentiviral expression of two independent shRNA constructs against HSD3B1 (shHSD3B1 #1 and shHSD3B1 #2) silences 3βHSD1 protein expression in LNCaP. The 3βHSD1 protein was quantitated and normalized to cells expressing nonsilencing lentiviral vector (shCTRL) and β-actin. (B) Silencing 3βHSD1(367T) blocks flux from [3H]-DHEA (100 nM) to AD as well as further downstream conversion to 5α-dione and DHT. Cells were treated with [3H]-DHEA in triplicate and steroids were quantitated with HPLC at the designated time points. (C) Inhibition of AR-regulated genes. Cells were treated with the indicated concentration of DHEA for 24 hours, and gene expression was assessed by qPCR and normalized to shCTRL-infected cells treated with vehicle and the RPLP0 housekeeping gene. (D) Silencing 3βHSD1(367T) inhibits in vitro growth. Cells were grown in the presence of 20 nM DHEA or vehicle and growth for each cell line is normalized to vehicle for each designated day. (E) 3βHSD1(367T) depletion blocks CRPC growth in LNCaP xenografts. Mice underwent surgical orchiectomy and DHEA pellet implantation concomitantly when xenograft tumors reached a threshold volume of 100 mm3. Fifteen mice were initiated in each cohort, 7, 8 and 10 mice in shCTRL, shHSD3B1 #1, and shHSD3B1 #2 groups, respectively, achieved a tumor volume of 100 mm3 in eugonadal mice, underwent orchiectomy and were included in the CRPC analysis. The number of days from orchiectomy to tumor volume ≥ 600 mm3 is shown. In the comparisons of shCTRL vs shHSD3B1 #1 and shHSD3B1 #2, P = 0.002 and 0.003, respectively, using a log rank test. (F) 3βHSD1(367T) protein is regained in CRPC tumors that grow from LNCaP expressing shHSD3B1 #1 and shHSD3B1 #2. Immunoblot for 3βHSD1 and β-actin were performed on protein from the indicated LNCaP CRPC tumors. Error bars in B, C and D represent the SD for experiments performed in triplicate.
3βHSD1(367T) is resistant to ubiquitination and degradation
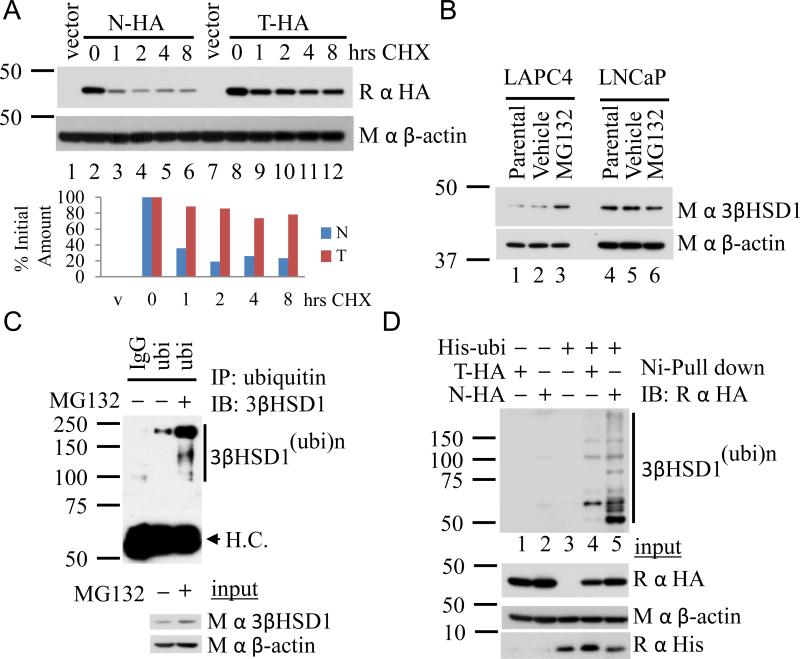
Endogenous expression of 3βHSD1(367T) appears to engender increased protein abundance compared to 3βHSD1(367N) expression (Fig 1E). To determine if the underlying mechanism is due to an alteration in protein degradation, wild-type (HSD3B1(N)-HA) and (HSD3B1(T)-HA) constructs were generated and transiently expressed, and protein levels were compared following inhibition of translation with cycloheximide (CHX) treatment. The 367 N → T mutation substantially increases protein half-life from 2.1 hours to 27 hours (Fig 4A). Similar experiments with an alternative prostate cancer cell line (Fig S3A) and with stable expression of lentiviral constructs confirm the longer half-life of 3βHSD1(367T) (Fig S3B). To determine whether increased degradation of wild-type protein is reversible with proteasome inhibition, cells were treated with MG132. Pharmacologic proteasome inhibition increases endogenous wild-type 3βHSD1(367N) in LAPC4 but not 3βHSD1(367T) in LNCaP (Fig 4B) and polyubiquitinated endogenous 3βHSD1(367N) accumulates with MG132 treatment in LAPC4 (Fig 4C). In contrast, polyubiquitinated endogenous 3βHSD1(367T) is not increased in LNCaP with MG132 treatment (Fig S3C). A direct comparison of ubiquitination between HA-tagged wild-type and mutant protein by Ni-agarose pull down demonstrates that 3βHSD1(367T) is resistant to polyubiquitination (Fig 4D), explaining decreased vulnerability to proteasome-mediated degradation and longer protein half-life.
Fig. 4.
Resistance to ubiquitination and proteosome-mediated degradation occurs with 3βHSD1(367T) which results in prolonged protein half-life. (A) 3βHSD1(367T) persists after inhibition of protein translation. LAPC4 cells were transiently transfected with constructs encoding for wild-type (N-HA) and (T-HA) protein and treated with cycloheximide (CHX) for the designated incubation times. Western blot with anti-HA antibody was performed, and signal was quantitated and normalized to time zero and β-actin. (B) Treatment with MG132 (10 μM; 8 hours) reverses 3βHSD1(367N) protein loss in LAPC4 and results in no 3βHSD1(367T) protein increase in LNCaP. (C) Proteosome inhibition with MG132 (10 μM; 8 hours) results in an increase in polyubiquitinated 3βHSD1(367N) protein in LAPC4 as evidenced by immunoprecipitation with an anti-ubiquitin antibody. (D) Loss of 3βHSD1(367T) vulnerability to proteosome-mediated degradation is explained by diminished susceptibility to ubiquitination. His-ubiquitin (His-ubi) was expressed with wild-type (N-HA) or (T-HA) protein in 293 cells, followed by pull down with Ni-agarose beads and anti-HA immunoblot.
AMFR binds 3βHSD1(367N) and is required for ubiquitination
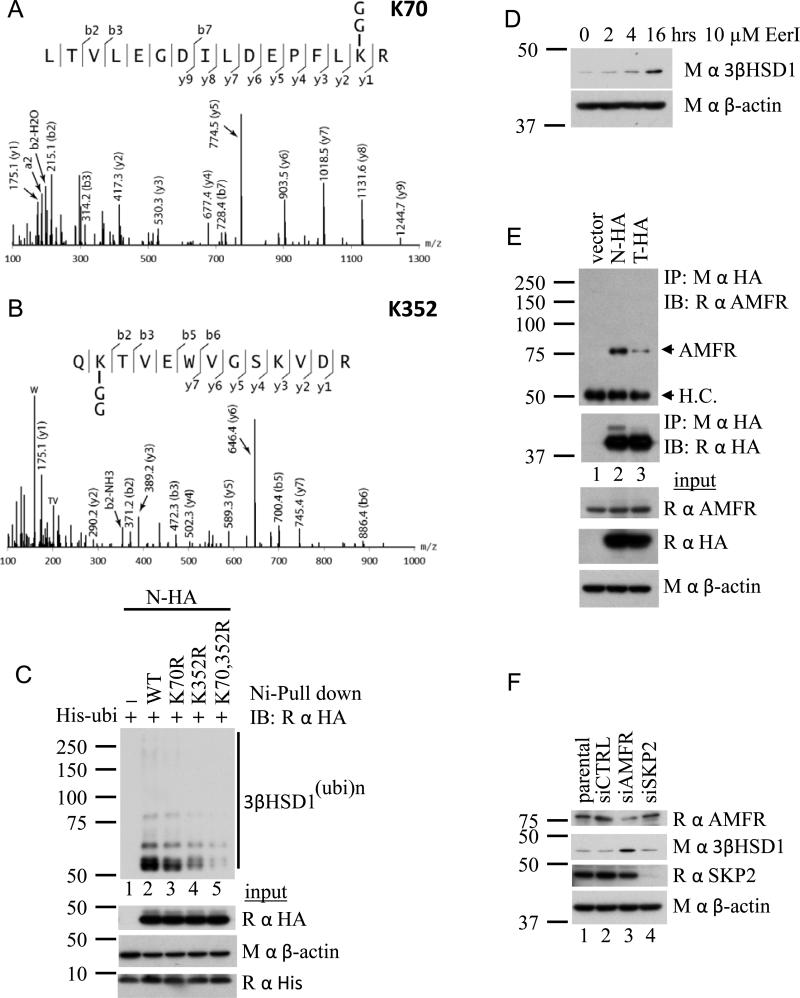
We employed mass spectrometry to determine the lysine residue(s) ubiquitinated on 3βHSD1(367N). Ubiquitination is detectable on both K70 (Fig 5A) and K352 (Fig 5B) of 3βHSD1(367N). The effects of K352R and K70R single mutations and a double mutation on ubiquitination were assessed by Ni-agarose pull down (Fig 5C). K352 appears to be a more critical site of ubiquitination than K70, and mutation of both sites decreases ubiquitination greater than either mutation alone. AMFR (autocrine mobility factor receptor, also known as gp78) is a membrane-anchored ubiquitin ligase that functions through the endoplasmic reticulum associated protein degradation (ERAD) pathway (Song et al., 2005). Eeyarestatin I (EerI) is a small molecule that inhibits protein degradation through the ERAD pathway (Wang et al., 2008; Wang et al., 2009). Endogenous 3βHSD1(367N) protein increases in LAPC4 cells with EerI treatment, suggesting that the ERAD pathway is required for 3βHSD1(367N) degradation (Fig 5D). Stable isotope labeling by amino acids in cell culture (SILAC) coupled with high-resolution mass spectrometry was employed to identify candidate ubiquitin ligases in an unbiased manner that preferentially associate with 3βHSD1(367N) (Ong et al., 2002). In this experiment, cells expressing 3βHSD1(367T)-HA and 3βHSD1(367N)-HA were grown in light and heavy media, respectively. AMFR was detected with a normalized protein ratio of 1.67 (derived from peptide ratios varying ≤17%) in a mixture of 3βHSD1(367N)-HA and 3βHSD1(367T)-HA immunoprecipitations mixed in a 1:1 ratio, indicating preferential physical association with 3βHSD1(367N) protein. Immunoprecipitation of 3βHSD1(367N)-HA and 3βHSD1(367T)-HA, followed by AMFR immunoblot confirms a preferential physical association of AMFR with 3βHSD1(367N) protein (Fig 5E). To assess the functional consequence of this interaction, AMFR was silenced using siRNA (Fig 5F). AMFR knockdown increases the abundance of 3βHSD1 protein, demonstrating the requirement of AMFR for 3βHSD1 degradation through the ERAD pathway. In contrast, silencing the alternative ubiquitin ligase SKP2 by siRNA has no detectable effect on 3βHSD1.
Fig. 5.
The ER-associated degradation (ERAD) pathway and AFMR regulate 3βHSD1 ubiquitination and degradation. (A, B) K70 and K352 ubiquitination on 3βHSD1(367N) is detectable by mass spectrometry. (C) K70, 352R mutant 3βHSD1(367N) is resistant to ubiquitination. K70R and K352R single and double mutant forms of N-HA were expressed with His-ubi in 293 cells, followed by pull down with Ni-agarose beads and anti-HA immunoblot. (D) Treatment with the ERAD inhibitor, Eeyarestatin I (EerI, 10μM), increases endogenous 3βHSD1 protein in LAPC4. (E) AMFR preferentially physically associates with wild-type protein (N-HA). Proteins were expressed in 293 cells, immunoprecipitated with anti-HA antibody, followed by immunoblot for AMFR. (F) Silencing the ubiquitin E3-ligase AMFR increases 3βHSD1 protein detected in LAPC4 cells. In contrast, genetically silencing the ubiquitin E3-ligase SKP2 has no detectable effect on 3βHSD1.
3βHSD1(367T) increases DHT synthesis
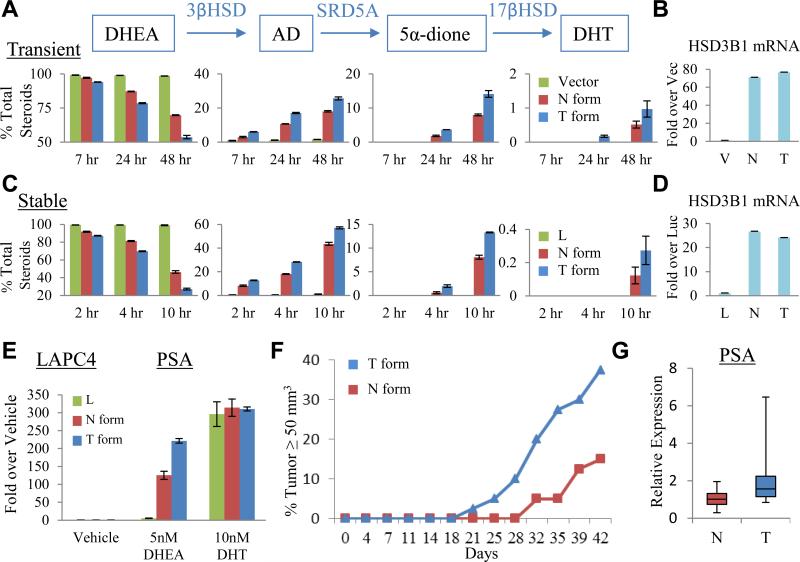
To determine if resistance to protein ubiquitination and degradation ascribed to 3βHSD1(367T) confers increased synthesis of DHT from precursor steroids, we expressed constructs that encode for 3βHSD1(367N), 3βHSD1(367T), or vector alone in LAPC4 cells, and assessed metabolic flux from [3H]-DHEA to downstream steroids. LAPC4 cells transiently transfected with the construct encoding for 3βHSD1(367T) exhibit increased flux from DHEA → AD → 5α-dione → DHT (Fig 6A). Equivalent expression of both transcripts was confirmed by qPCR (Fig 6B). Stable lentiviral expression of 3βHSD1(367T) similarly confers increased flux from DHEA → AD → 5α-dione → DHT (Fig 6C) with transcript expression comparable to wild-type (Fig 6D). Finally, we determined that the 3βHSD1(367T) phenotype that accelerates flux from DHEA to DHT amplifies the response of androgen-regulated gene expression (Fig 6E) and hastens the time to the development of CRPC xenograft tumors (Fig 6F) in orchiectomized mice supplemented with DHEA to mimic human adrenal physiology. 3βHSD1(367T) tumors express higher levels of PSA transcript compared to 3βHSD1(367N) tumors, suggesting the presence of higher sustained DHT concentrations generated in 3βHSD1(367T) tumors (Fig 6G). Together, these findings support a mechanism favoring genetic selection for the allele encoding 3βHSD1(367T) in the setting of androgen depletion.
Fig. 6.
3βHSD1(367T) increases metabolic flux from DHEA to DHT and elicits CRPC. (A) Transient expression of 3βHSD1(367T) (T, blue bars) leads to increased conversion from DHEA to AD and downstream steroids compared with 3βHSD1(367N) (N, red bars). LAPC4 cells were transfected with the indicated plasmid, treated with CHX, and cultured with [3H]-DHEA (100 nM); steroids were extracted and measured by HPLC at the designated time points (p-value = 0.023 for the difference in DHT synthesis by the N and T forms using by Student's t-test). (B) Transient transfection results in equivalent expression of both transcripts by qPCR. (C) Stable expression demonstrates increased activity of 3βHSD1(367T). Lentiviral constructs expressing luciferase (L), wild-type (N), or (T), were stably expressed (without CHX treatment) and flux from [3H]-DHEA to DHT was assessed, as described previously (p-value = 0.015 for the difference in DHT synthesis by the N and T forms using Student's t-test). (D) Expression of both enzyme transcripts by qPCR is comparable. (E) Increased flux from DHEA to DHT with stable expression of 3βHSD1(367T) leads to amplified expression of PSA in LAPC4. Cells stably expressing the designated constructs were treated with the indicated steroids for 48 hours. PSA expression induced by the DHT positive control is equivalent among the three cell populations. For B, D and E, expression is normalized to RPLP0 and vector, luciferase, or vehicle controls. Error bars represent the SD for experiments performed in triplicate. (F) Development of CRPC occurs more rapidly in LAPC4 xenografts stably expressing 3βHSD1(367T) as compared with 3βHSD1(367N). Time from subcutaneous injection of cells in each flank to tumor size = 50 mm3 is shown for each tumor that developed in a mouse flank (n = 40 mouse flanks in each group). P=0.017 for the comparison using a log rank test. (G) PSA expression is higher in CRPC tumors expressing 3βHSD1(367T) compared with 3βHSD1(367N) (p-value = 0.015 by Student's t-test). Expression is normalized to RPLP0. Bars represent the upper and lower quartiles of individual tumor values. See also Figure S3.
Discussion
A major mechanism of resistance to frontline gonadal T depletion (or castration) therapy is an acquired metabolic capability, which allows CRPC tumors to sustain sufficient DHT concentrations for AR stimulation and tumor progression. This study is the first to identify a gain-of-function mutation in the steroidogenic machinery that increases flux to DHT. Notably, whether utilizing the major adrenal pathway or possibly de novo steroidogenesis from cholesterol, 3β-hydroxl oxidation to 3-keto and Δ5→4 isomerization by 3βHSD enzymatic activity is required for all pathways culminating in T and/or DHT synthesis (Evaul et al., 2010). Adrenal DHEA and DHEA-sulfate are typically present in abundant concentrations in human serum. In the context of intratumoral 3βHSD1(367N) expression, the clinical response to gonadal T depletion probably occurs in part due to the limited contribution of adrenal precursors to intratumoral DHT. Augmented 3βHSD activity occurring through increased protein abundance with 3βHSD1(367T) would therefore serve to open the floodgates on a proximal and otherwise rate-limiting step for the synthesis of DHT, resulting in the development of CRPC. Notably, in the setting of heterozygous inheritance, 3βHSD1 protein expression is markedly higher in tumors that have lost the wild-type HSD3B1(1245A) allele compared to tumors that retain the wild-type sequence (Fig 2C). This finding might occur because expression and co-localization of the wild-type 3βHSD1(367N) protein reinstates mutant 3βHSD1(367T) ubiquitination and subsequent degradation via dimerization or oligomerization. Nonetheless, engineered 3βHSD1(367T) expression engenders increased flux to DHT and development of CRPC despite endogenous 3βHSD1(367N) expression (Fig 6). Therefore, the transition from sole 3βHSD1(367N) expression to mixed expression to dominant 3βHSD1(367T) expression probably represents a stepwise selection for an increased capacity for DHT synthesis.
The population frequency of the HSD3B1(1245C) allele is approximately 22% but appears to vary widely by ethnicity (UCSC). Other studies suggest that the HSD3B1(1245C) allele may raise aldosterone levels and increase the risk of essential hypertension (Shimodaira et al., 2010). This is probably attributable to increased 3βHSD enzyme activity, which is required for aldosterone synthesis, although aldosterone is generally thought to require 3βHSD2. Interestingly, this phenotype appears to be more severe with homozygous HSD3B1(1245C). The observation of extremely high aldosterone with homozygous HSD3B1(1245C) is consistent with higher enzymatic activity and stepwise selection for sole 3βHSD1(367T) expression that occurs in CRPC. HSD3B1(1245C) has no consistent effect on risk of localized prostate cancer (Chang et al., 2002; Cunningham et al., 2007; Thomas et al., 2008).
Although abiraterone potently inhibits androgen synthesis, clinical studies of urinary androgen metabolites in patients with CRPC treated with this drug have demonstrated that the block is incomplete and that the synthesis of residual androgen precursors persists (Attard et al., 2012). This finding raises the possibility that tumor mechanisms that augment androgen synthesis from limited precursor steroids by increasing flux to DHT might contribute to abiraterone resistance (Chang and Sharifi, 2012). Our data demonstrating the selection and expression of 3βHSD1(367T) in a xenograft model of abiraterone resistance suggest a genetic mechanism for clinical resistance to abiraterone and that pharmacologic inhibition of 3βHSD1 might be a viable therapeutic strategy to overcome this resistance against tumors expressing the mutant enzyme. Despite the potent activity of the AR antagonist enzalutamide, its affinity for the ligand-binding domain of AR is lower than the affinity of DHT (Tran et al., 2009). Increased metabolic flux from steroid precursors to DHT by 3βHSD1(367T) may therefore conceivably tip the scales in the favor of DHT and lead to enzalutamide resistance as well. The contribution of 3βHSD1(367T) in clinical resistance to abiraterone and enzalutamide, however, remain to be determined.
The past decade has brought to the fore the development of molecularly targeted therapies that are matched to specific disease-driving enzyme mutations present in a given patient. These advances come mainly in the form of tyrosine kinase inhibitors that target gain-of-function mutations in these signaling enzymes. These include the examples of EGF receptor inhibitors matched with tumors harboring mutant EGF receptor in non-small cell lung cancer and BRAF inhibitors for melanomas that are driven by BRAF mutations (Chapman et al., 2011; Kobayashi et al., 2005; Lynch et al., 2004). In contrast, no examples of drug targeting based on enzyme mutations exist in the standard of care for metastatic CRPC. While our demonstration is in a gain-of-function in a metabolic enzyme, rather than a signaling enzyme, we believe the underlying principle is the same, and our findings expose the opportunity for matching a mutant disease-driving enzyme biomarker with its cognate pharmacologic inhibitor.
Experimental Procedures
Steroid Metabolism Experiments
Steroid metabolism experiments were performed 12 hours after seeding cells by treatment with 1 mL serum-free medium containing [3H]-labeled DHEA (100 nM, 300,000-600,000 cpm; PerkinElmer). Aliquots of medium were collected for up to 48 hours, treated with β-glucuronidase (1000 units; Sigma-Aldrich) at 65°C for 4 hours. Deconjugated steroids were extracted, evaporated under nitrogen stream, dissolved in 50% methanol, injected on a Breeze 1525 system equipped with model 717 plus autoinjector (Waters Corp.) and steroid metabolites were separated on a Luna 150 × 3 mm, 3.0 μM C18 reverse-phase column (Phenomenex). The column effluent was mixed with Liquiscint scintillation cocktail (National Diagnostics) and analyzed by a β-RAM model 3 in-line radioactivity detector (IN/US Systems). Steroid metabolism experiments with transient enzyme expression were performed with pCMV5-HSD3B1 (367N and 367T) constructs 24 hours after transfection and 12 hours after treatment with 25 μM cycloheximide (CHX). Steroid metabolism experiments with stable enzyme expression were performed after lentiviral infection with pLVX-Tight-Puro vector. Human tissues were obtained using IRB approved protocols at UT Southwestern and the University of Washington rapid autopsy program. Lentiviral constructs were made from miR30-styled shRNA sequences and cloned into the pGIPZ vector and infected cells expressing the constructs were selected with 2 μg/mL puromycin. Gene expression was performed by qPCR using the iTaq SYBR Green Supermix with the ROX kit (Bio-Rad) in an ABI-7500 Real-Time PCR machine (Applied Biosystems). Protein half-life was determined after transient transfection with pCMX-HSD3B1-HA (367N and 367T) plasmids, followed in 24 hours with 25 μM CHX in serum-free medium containing 100 nM DHEA. Cells stably expressing HA-tagged HSD3B1 (367N and 367T) in pLVX-Tight-Puro were used to determine protein half-life 24 hours after induction of protein expression with 2 ng/mL doxycycline and treatment with CHX.
For details on all other experiments including cell line and human tissue analyses, xenograft studies, gene expression studies, mass spectrometry and other biochemical experiments, please refer to the Extended Experimental Procedures.
Supplementary Material
Article Highlights.
3βHSD1 catalyzes a rate-limiting step for DHT synthesis in CRPC
Selection for N367T mutant 3βHSD1 occurs in human CRPC tumors
The N367T 3βHSD1 mutation confers resistance to ubiquitination and degradation
Mutant 3βHSD1 protein accumulates, increasing DHT synthesis and causing CRPC
One Sentence Summary.
A gain-of-function in HSD3B1 increases DHT synthesis, eliciting resistance to androgen deprivation therapy in prostate cancer.
Acknowledgements
We thank Ralph Deberardinis, Mike Brown, Kevin Courtney, George DeMartino and Eugene Frenkel for helpful comments, Russell DeBose-Boyd for the anti-AFMR antibody, J.T. Hsieh for the anti-SKP2 antibody, Cheng-Ming Chiang for the anti-His antibody and Actinomycin D, David Trudgian for assistance with protein mass spectrometry and An Jia and Chul Ahn for assistance with statistical analysis. This publication has been funded in part by a Howard Hughes Medical Institute Physician-Scientist Early Career Award (NS), the Prostate Cancer Foundation (NS, RV, PSN), an American Cancer Society Research Scholar Award (12-038-01-CCE) (NS), grant PC080193 from the U.S. Army Medical Research and Materiel Command (NS), 1R01CA168899 (NS) and 1R01CA172382-01 (NS). The acquisition of metastatic tumors through the UW rapid autopsy program was funded in part by grants PO1-CA85859 (RV, PSN), PC093509 (PSN) and P50CA097186 (RV, PSN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- UCSC Genome Browser. http://genome.ucsc.edu/cgibin/hgc?hgsid=300408599&c=chr1&o=120057245&t=120057246&g=snp135Common&i=rs1047303.
- Attard G, Cooper CS, de Bono JS. Steroid hormone receptors in prostate cancer: a hard habit to break? Cancer Cell. 2009;16:458–462. doi: 10.1016/j.ccr.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Attard G, Reid AH, Auchus RJ, Hughes BA, Cassidy AM, Thompson E, Oommen NB, Folkerd E, Dowsett M, Arlt W, et al. Clinical and Biochemical Consequences of CYP17A1 Inhibition with Abiraterone Given with and without Exogenous Glucocorticoids in Castrate Men with Advanced Prostate Cancer. J Clin Endocrinol Metab. 2012;97:507–516. doi: 10.1210/jc.2011-2189. [DOI] [PubMed] [Google Scholar]
- Barrie SE, Potter GA, Goddard PM, Haynes BP, Dowsett M, Jarman M. Pharmacology of novel steroidal inhibitors of cytochrome P450(17) alpha (17 alpha-hydroxylase/C17-20 lyase). J Steroid Biochem Mol Biol. 1994;50:267–273. doi: 10.1016/0960-0760(94)90131-7. [DOI] [PubMed] [Google Scholar]
- Chang BL, Zheng SL, Hawkins GA, Isaacs SD, Wiley KE, Turner A, Carpten JD, Bleecker ER, Walsh PC, Trent JM, et al. Joint effect of HSD3B1 and HSD3B2 genes is associated with hereditary and sporadic prostate cancer susceptibility. Cancer Res. 2002;62:1784–1789. [PubMed] [Google Scholar]
- Chang KH, Li R, Papari-Zareei M, Watumull L, Zhao YD, Auchus RJ, Sharifi N. Dihydrotestosterone synthesis bypasses testosterone to drive castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2011;108:13728–13733. doi: 10.1073/pnas.1107898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KH, Sharifi N. Prostate cancer-from steroid transformations to clinical translation. Nat Rev Urol. 2012 doi: 10.1038/nrurol.2012.175. [DOI] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham JM, Hebbring SJ, McDonnell SK, Cicek MS, Christensen GB, Wang L, Jacobsen SJ, Cerhan JR, Blute ML, Schaid DJ, et al. Evaluation of genetic variations in the androgen and estrogen metabolic pathways as risk factors for sporadic and familial prostate cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:969–978. doi: 10.1158/1055-9965.EPI-06-0767. [DOI] [PubMed] [Google Scholar]
- de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB, Jr., Saad F, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evaul K, Li R, Papari-Zareei M, Auchus RJ, Sharifi N. 3β-hydroxysteroid dehydrogenase is a possible pharmacological target in the treatment of castration-resistant prostate cancer. Endocrinology. 2010;151:3514–3520. doi: 10.1210/en.2010-0138. [DOI] [PubMed] [Google Scholar]
- Geller J, Albert J, Loza D, Geller S, Stoeltzing W, de la Vega D. DHT concentrations in human prostate cancer tissue. J Clin Endocrinol Metab. 1978;46:440–444. doi: 10.1210/jcem-46-3-440. [DOI] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen KE, Penning TM. Partners in crime: deregulation of AR activity and androgen synthesis in prostate cancer. Trends Endocrinol Metab. 2010;21:315–324. doi: 10.1016/j.tem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Li R, Evaul K, Sharma KK, Chang K-H, Yoshimoto J, Liu J, Auchus RJ, Sharifi N. Abiraterone Inhibits 3β-Hydroxysteroid Dehydrogenase: A Rationale for Increasing Drug Exposure in Castration-Resistant Prostate Cancer. Clinical Cancer Research. 2012;18:3571–3579. doi: 10.1158/1078-0432.CCR-12-0908. [DOI] [PubMed] [Google Scholar]
- Lorence MC, Murry BA, Trant JM, Mason JI. Human 3 beta-hydroxysteroid dehydrogenase/delta 5----4isomerase from placenta: expression in nonsteroidogenic cells of a protein that catalyzes the dehydrogenation/isomerization of C21 and C19 steroids. Endocrinology. 1990;126:2493–2498. doi: 10.1210/endo-126-5-2493. [DOI] [PubMed] [Google Scholar]
- Luu-The V, Belanger A, Labrie F. Androgen biosynthetic pathways in the human prostate. Best Pract Res Clin Endocrinol Metab. 2008;22:207–221. doi: 10.1016/j.beem.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Montgomery RB, Mostaghel EA, Vessella R, Hess DL, Kalhorn TF, Higano CS, True LD, Nelson PS. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68:4447–4454. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Penning TM. New frontiers in androgen biosynthesis and metabolism. Curr Opin Endocrinol Diabetes Obes. 2010;17:233–239. doi: 10.1097/MED.0b013e3283381a31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller MD, de Wit R, Mulders P, Chi KN, Shore ND, et al. Increased Survival with Enzalutamide in Prostate Cancer after Chemotherapy. N Engl J Med. 2012 doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- Sharifi N. Minireview: androgen metabolism in castration-resistant prostate cancer. Mol Endocrinol. 2013;27:708–714. doi: 10.1210/me.2013-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi N, Gulley JL, Dahut WL. Androgen deprivation therapy for prostate cancer. JAMA. 2005;294:238–244. doi: 10.1001/jama.294.2.238. [DOI] [PubMed] [Google Scholar]
- Shimodaira M, Nakayama T, Sato N, Aoi N, Sato M, Izumi Y, Soma M, Matsumoto K. Association of HSD3B1 and HSD3B2 gene polymorphisms with essential hypertension, aldosterone level, and left ventricular structure. Eur J Endocrinol. 2010;163:671–680. doi: 10.1530/EJE-10-0428. [DOI] [PubMed] [Google Scholar]
- Simard J, Ricketts ML, Gingras S, Soucy P, Feltus FA, Melner MH. Molecular biology of the 3beta-hydroxysteroid dehydrogenase/delta5-delta4 isomerase gene family. Endocr Rev. 2005;26:525–582. doi: 10.1210/er.2002-0050. [DOI] [PubMed] [Google Scholar]
- Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Titus MA, Schell MJ, Lih FB, Tomer KB, Mohler JL. Testosterone and dihydrotestosterone tissue levels in recurrent prostate cancer. Clin Cancer Res. 2005;11:4653–4657. doi: 10.1158/1078-0432.CCR-05-0525. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–790. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li L, Ye Y. Inhibition of p97-dependent protein degradation by Eeyarestatin I. J Biol Chem. 2008;283:7445–7454. doi: 10.1074/jbc.M708347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Mora-Jensen H, Weniger MA, Perez-Galan P, Wolford C, Hai T, Ron D, Chen W, Trenkle W, Wiestner A, et al. ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells. Proc Natl Acad Sci U S A. 2009;106:2200–2205. doi: 10.1073/pnas.0807611106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Balk SP. Mechanisms mediating androgen receptor reactivation after castration. Urol Oncol. 2009;27:36–41. doi: 10.1016/j.urolonc.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.