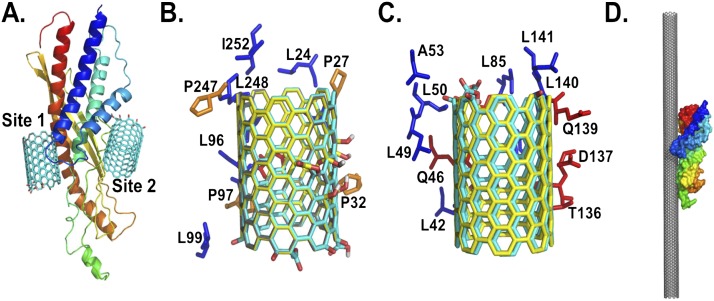
Figure 7.
Structural details of the predicted binding sites of nonoxidized and oxidized SWCNTs on SPLUNC1. (A) Structural model of SPLUNC1, along with the two predicted binding sites 1 and 2, with oxidized SWCNTs at the ends. The residues that are within close proximity of SPLUNC1 (within 4 Å) stabilize the binding sites. Amino-acid compositions of the predicted Site 1 (B) and (C) Site 2 of SPLUNC1 interact with nonoxidized SWCNTs (gray), oxidize SWCNTs at the ends (cyan), and (D) oxidize SWCNTs in the middle (yellow). The predicted binding orientation of SPLUNC1 to a SWCNT surface is demonstrated in a 25-nm length. The structure of SPLUNC1 in A and D is rendered as cartoon and surface, respectively, and is colored as a rainbow from the N-terminus to the C-terminus. The proline, hydrophobic, and polar residues in B and C are colored in orange, blue, and red, respectively, and are rendered as sticks. A, alanine; D, aspartic acid; I, isoleucine; L, leucine; P, proline; Q, glutamine; T, threonine.