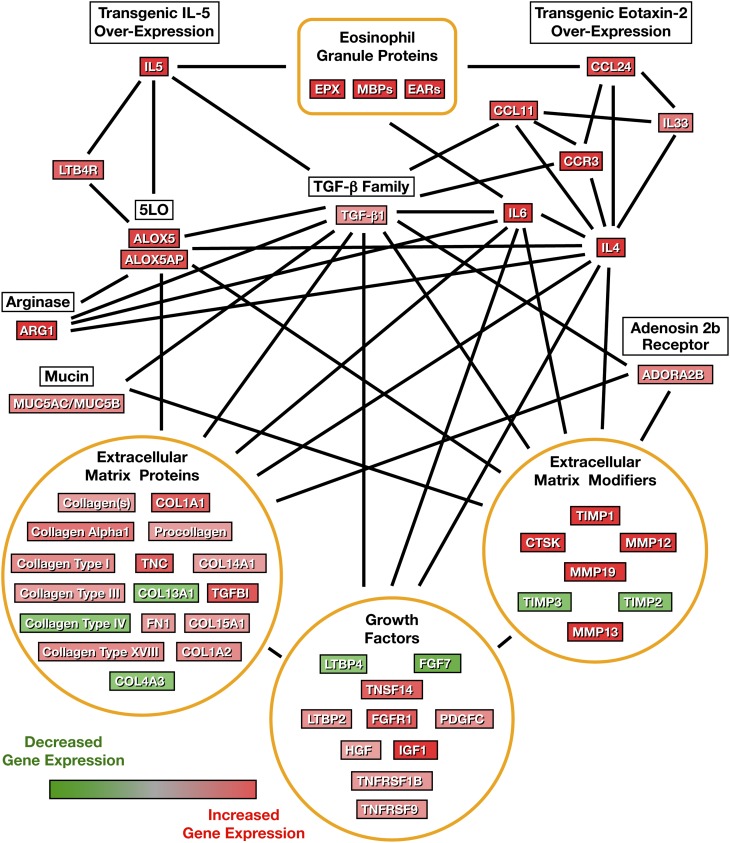
Figure 2.
Compilation of airway molecular pathways derived from assessments of differential gene expression in the lungs of eosinophil-sufficient I5/hE2 versus eosinophilless PHIL/I5/hE2 mice. Ingenuity Pathway Analysis of the transcript array profiles derived from eosinophil-sufficient I5/hE2 versus eosinophilless PHIL/I5/hE2 mice was used in conjunction with previously published studies to identify four molecular networks (lipid metabolism, airway fibrosis and remodeling, airway hyperresponsiveness [AHR], and asthma [Table E1]) relevant to chronic pulmonary diseases. These pathways were then combined to create a larger network scheme of gene expression linked with the eosinophil-mediated inflammation associated with I5/hE2 mice.