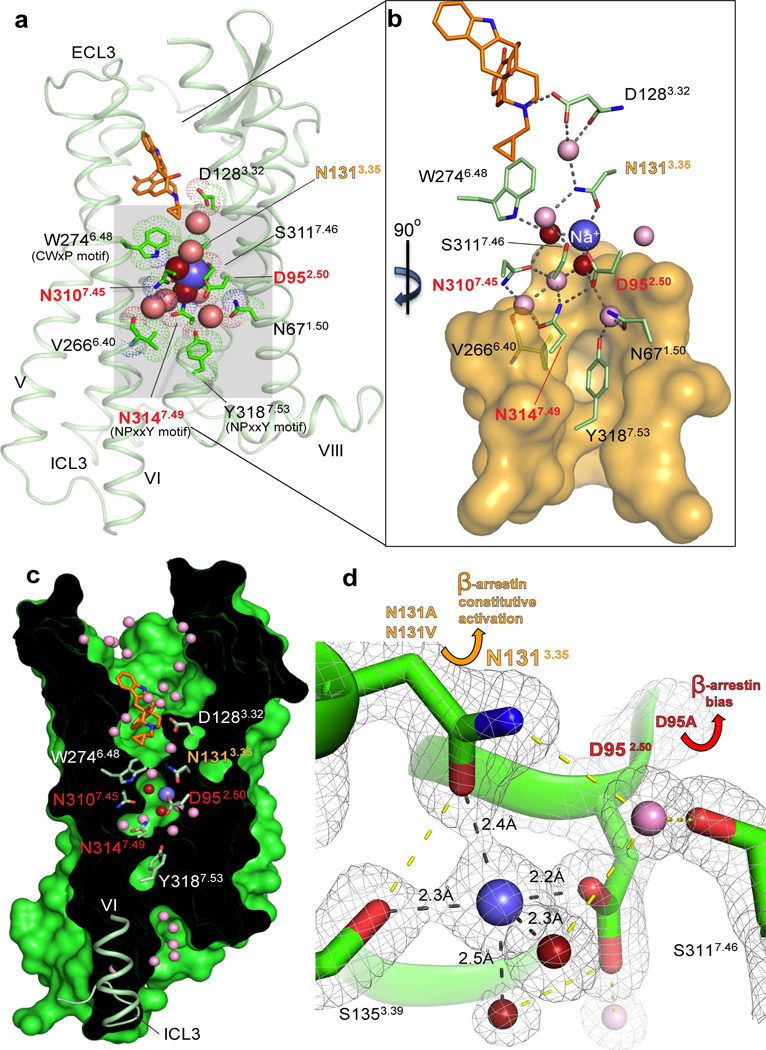
Fig. 1. Interactions in the 7TM core of BRIL-δOR(ΔN/ΔC)-naltrindole.
(a) BRIL-δOR(ΔN/ΔC)-naltrindole structure (light green, BRIL fusion is omitted) and residues in the allosteric sodium site (green sticks). Sodium is shown as a blue sphere; red and pink spheres are waters in the first and second coordination shells, respectively. Naltrindole is shown as orange sticks. (b) Interactions in the sodium site. Orange transparent surface depicts hydrophobic residues below the allosteric sodium site. Hydrogen bonds are shown as grey dotted lines. (c) ‘Sliced’ surface representation of BRIL-δOR(ΔN/ΔC)-naltrindole showing the continuous pathway connectivity between the orthosteric and allosteric sodium site. Receptor surface and interior are colored green and black, respectively. (d) 2mFo- DFcelectron density map (grey mesh) contoured at 2 σ around residues, waters and sodium in the allosteric site. Hydrogen bonds in the first sodium ion coordination shell are shown as black dotted lines; all other hydrogen bonds are shown as yellow dotted lines. Yellow arrow indicates the increased β-arrestin constitutive activity of Asn1313.35Ala and Asn1313.35Val mutants. Red arrow indicates the potent β-arrestin biased activation and concomitant abrogation of Gi signaling in response to the ligand naltrindole in the Asp952.50Ala mutant. The “efficacy switch” residues that resulted in β-arrestin bias when mutated are highlighted in orange (Asn1313.35) and red (Asp952.50, Asn3107.45 and Asn3147.49) in panels a-d.