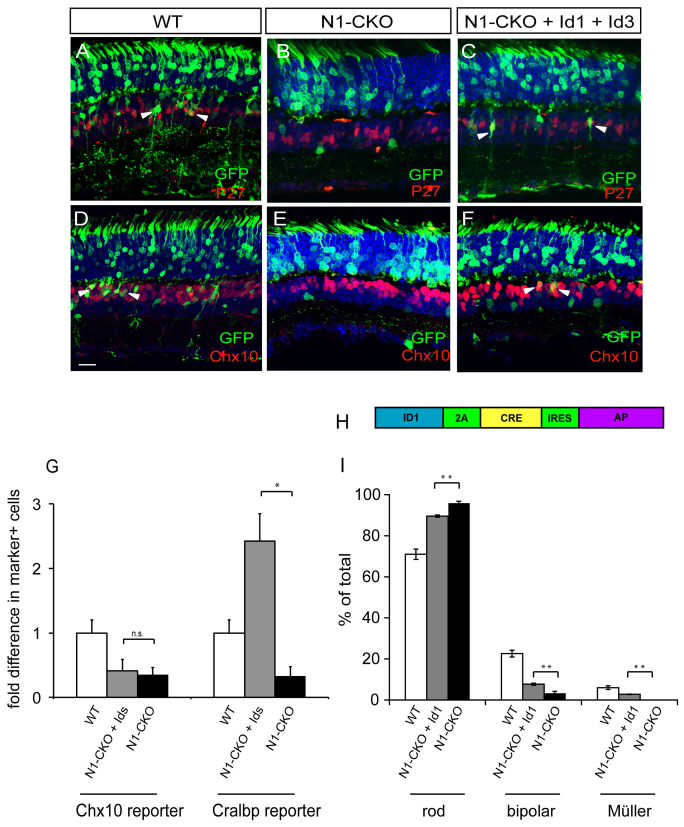
Fig. 5.
Misexpression of Id1 and Id3 in N1-CKO retinas. (A-F) Notch1fl/fl retinas were electroporated in vivo at P0 with CAG:GFP, or CAG:Cre and CALNL-GFP, or CAG:Cre, CALNL-GFP, CAG:Id1 and CAG:Id3. The fates of the electroporated cells were analyzed in the mature retina (after P14) using morphology, location and immunohistochemistry. Müller glial cells express p27 and bipolar cells express high levels of Chx10 (red). (A,D) Electroporation of CAG:GFP alone resulted in GFP+ photoreceptors, interneurons and Müller glial cells, some of which were positive for p27 or Chx10 (arrowheads indicate examples). (B,E) Electroporation of CAG:Cre and CALNL-GFP into Notch1fl/fl retinas resulted in GFP+ photoreceptors and some amacrine cells, none of which was positive for p27 or Chx10. (C,F) Electroporation of CAG:Cre, CALNL-GFP, CAG:Id1 and CAG:Id3 resulted in GFP+ photoreceptors, interneurons and Müller glial cells, some of which were positive for p27 and Chx10 (arrowheads demarcate examples). Scale bar: 50 μm. (G) Combinations of plasmids encoding CAG:GFP or CAG:Cre and CALNL-GFP, cell type-specific reporters (Chx10:tdTomato to mark bipolar cells and Cralbp:dsRed to mark Müller glial cells), and Id factors were co-electroporated in vitro into P0 Notch1fl/fl retinas. Retinas were cultured for 11-12 days and dissociated to single cells. FACS was used to determine the percentage of GFP+ cells that were dsRed+ or tdTomato+ for each condition. Fold difference is the percentage of GFP+marker+ cells in N1-CKO conditions, or induced by Id1+Id3 misexpression in N1-CKO conditions, relative to wild type. n=5 retinas per condition. *P<0.05; n.s., P>0.05. (H) The LIA retrovirus was engineered to express Id1, Cre and AP. Notch1fl/fl retinas were infected with LIA-Id1-2A-Cre-IRES-AP at P3 and the fate of one-cell clones was assessed after P21 by histochemical staining. (I) Quantification of cell types found in BAG (WT), LIA-Cre (N1-CKO) (as described in Fig. 1) and LIA-Id1-2A-Cre (N1-CKO+Id1) one-cell clones. n=3 retinas per condition, 464 BAG one-cell clones, 746 LIA-Id2-2A-Cre one-cell clones. **P<0.01.