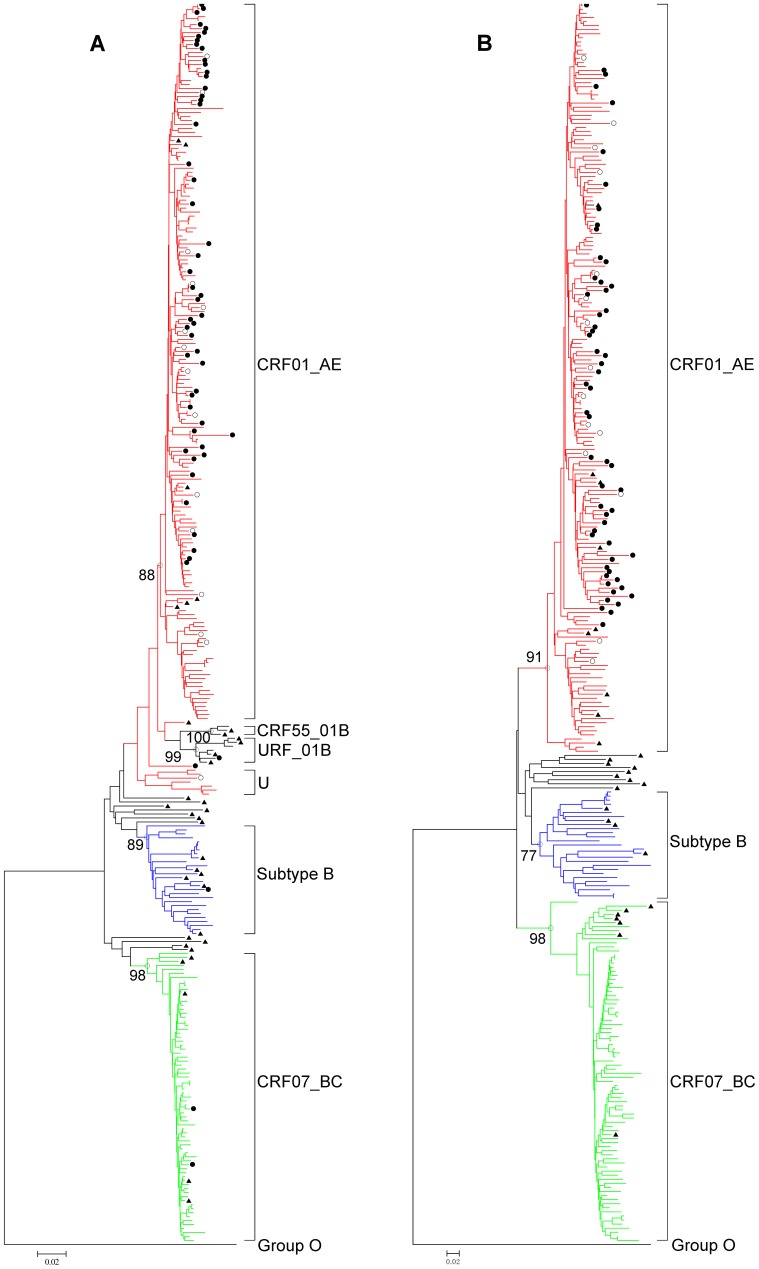
Figure 1. Phylogenetic tree analysis of HIV-1 env and pol gene sequences among MSM with recent infections in Shanghai.
The phylogenetic trees were constructed using neighbor-joining methods (Mega 5.0) based on pol (A) and env (B) sequence regions. The bootstrap values of 1000 replicates above 75% are labeled on the major clusters nodes. The CXCR4-tropic strains determined with algorithm I were indicated by both solid and open circles, whereas the CXCR4-tropic strains determined with algorithm II were only indicated by solid circles. CRF01_AE sequences are marked in red, CFR07_BC sequences are marked in green, and subtype B/B’ sequences are marked in blue. U stands for unidentified subtypes/recombinants. The subtype reference sequences from the Los Alamos HIV sequence database (http://hiv-web.lanl.gov/content/index) were indicated by solid triangles. Trees were rooted using group O as a out group.