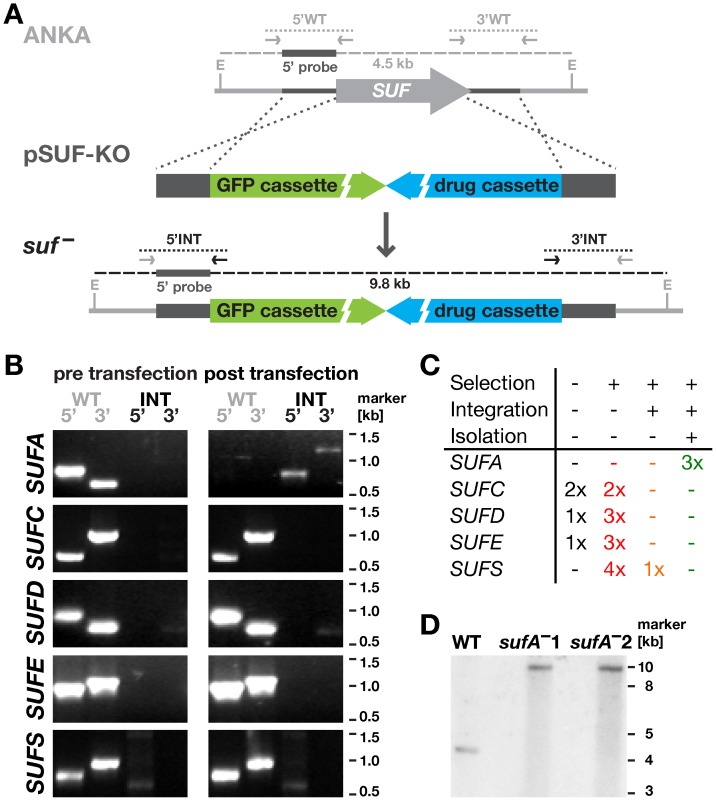
Figure 1. Systematic gene targeting of Plasmodium berghei SUF genes.
(A) Replacement strategy to delete the five nuclear-encoded PbSUF genes. The respective ANKA strain wild type (WT) SUF loci were targeted with replacement plasmids (pKO) containing upstream 5′ and downstream 3′ regions (dark gray bars) flanking the open reading frames (light gray arrow), a high-expressing GFP cassette (green), and the hDHFR-yFcu drug-selectable cassette (blue). Integration-specific (5′INT and 3′INT) and wild type-specific (5′WT and 3′WT) primer combinations (Table S1) are indicated by arrows; expected PCR fragments by dotted lines. The probe used for Southern blot analysis of the two isogenic sufA – parasites lines corresponds to the 5′ integration sequence and hybridized to EcoRI (E) restriction-digested gDNA; expected fragments and their sizes are indicated by gray dashed lines. (B) Representative diagnostic PCR results of the SUF loci of WT ANKA (pre transfection) and drug-selected (post transfection) parasites are shown. For SUFA, diagnostic PCR of isogenic gene deletion parasites confirms successful integration and absence of WT parasite contamination. (C) Overview of all transfection experiments summarizing the number of times no pyrimethamine-resistant parasites were selected (black), selection of pyrimethamine-resistant parasites was achieved (red), integration-specific PCR demonstrated targeted deletion of the SUF gene (orange), and isolation of WT-free, isogenic recombinant parasites (green). (D) Southern blot analysis of two isogenic sufA – parasite lines reveals the expected size shifts.