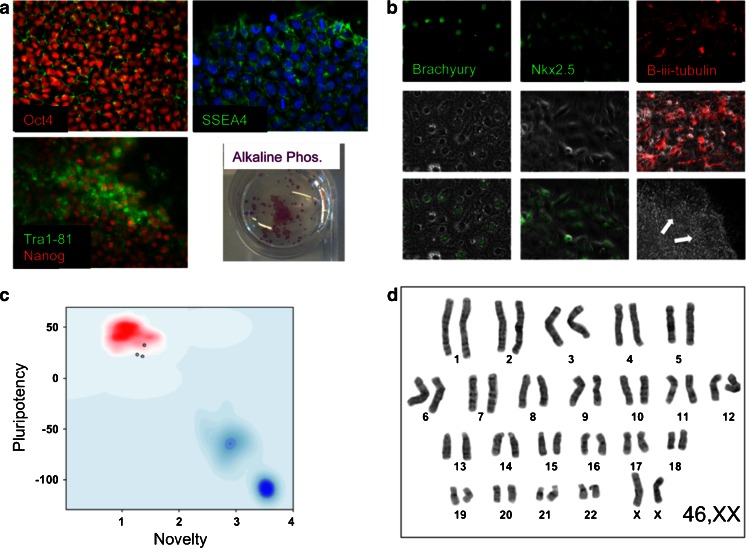
Fig. 2.
Representative images for the validation of iPSC pluripotency and quality control. a Undifferentiated iPSCs were probed with primary antibodies to Oct4, Nanog, SSEA4 and Tra1-81 and assessed for alkaline phosphatase enzymatic activity. b To assess the capacity of the iPS lines to spontaneously differentiate into the three embryonic lineages, EBs were generated in vitro for 2 days and then plated onto gelatin-coated plates. The cultures were grown for a further 12 days before fixing and staining for mesodermal, endodermal and neuroectodermal markers Brachyury, Nkx2.5 and β-iii-tubulin, respectively. c For the assessment of pluripotency using ‘PluriTest’, total RNA was extracted from iPS cells and genome wide gene expression analysis was performed on Illumina HT12v4 beadchips at the Genome Centre, Queen Mary, University of London. For Pluritest analysis, intensity data files were uploaded at http://www.pluritest.org and analysis performed according to Muller et al. (2011). d Karyotypes were generated from 70–80 % confluent cells treated with 100 ng/ml colcemid (Life Technologies) for 3 h and subjected to hypotonic lysis in 0.075 M potassium chloride for 10 min at 37 °C. Samples were then fixed in methanol/glacial acetic acid (ratio 3:1) and stained with Giemsa on glass slides for analysis. Twenty metaphase cells were counted for each line and four cells analysed in detail