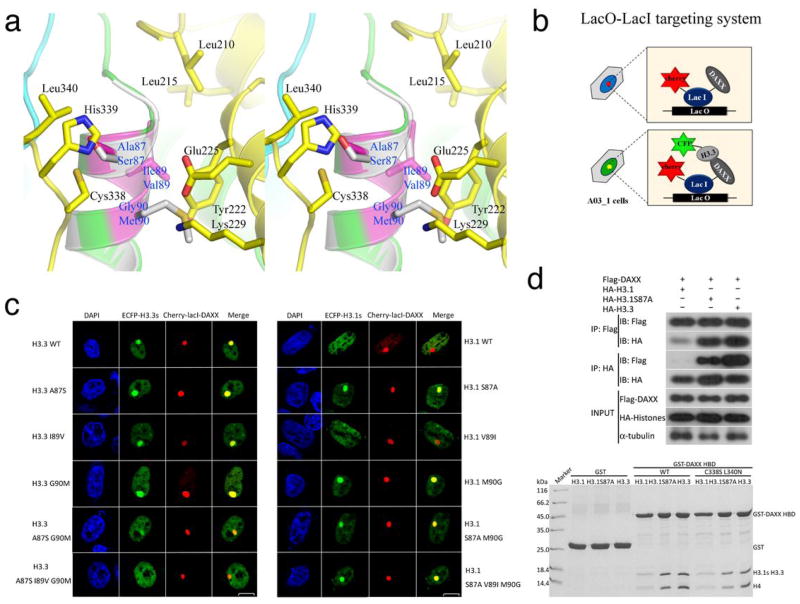
Figure 3.
H3.3 residues responsible for DAXX binding specificity. (A) A stereo view of H3.3-specific interactions with DAXX HBD. The three H3.3-specific residues are highlighted in magenta, and the interacting DAXX residues are shown (DAXX-yellow; H3.3-green or magenta; H4-cyan). Helix α2 of a canonical H3 (gray; PDB 1KX5) is superimposed and the three residues different from H3.3 are shown for comparison. (B) A schematic diagram showing the lac operator-repressor (LacO-LacI) targeting system for assaying DAXX-H3.3 interactions. (C) Co-localization of DAXX with H3.3, H3.1 and indicated mutants. (D) Top panel: co-IP of full-length Flag-tagged DAXX with HA-tagged histones H3.1 and H3.3. IPs were analyzed by western blotting with indicated antibodies. Bottom panel: GST pull-down of wild-type or the C338S L340N double mutant of DAXX HBD with the H3.1-H4, H3.1(S87A)-H4, or H3.3-H4 complexes as indicated. The pull-downs were analyzed by Coomassie-stained SDS-PAGE.