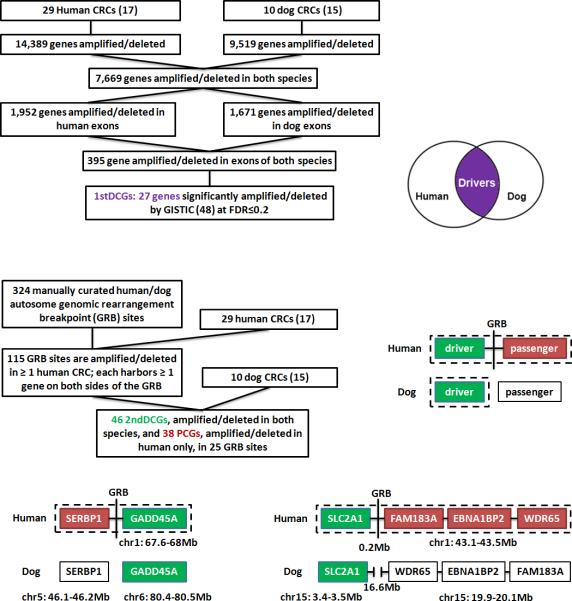
Fig. 1. CRC driver-passenger distinction via human-dog comparison.
The top portion illustrates how 1stDCGs (27 total), shown in purple and with at least one of their exons significantly amplified/deleted in both species, were identified autosome-wide (see the main text and Supplementary Table s1 for details). The middle section indicates how 2ndDCGs (46 total), shown in green, and PCGs (38 total), shown in red, were identified from the human-dog GRB sites in the human genome (see the main text for criteria for DCG-PCG distinction; Supplementary Tables s1-s3 and Fig. s1 for details). The bottom area shows examples of DCG-PCG pairs identified from two GRB sites in the human genome that harbor a human-dog translocation breakpoint (left) or a human-dog inversion breakpoint (right). Genes in each example are clustered in the human genome, but are either located on different chromosomes (right), or far apart (0.2 Mb for the human versus 16.6Mb for the dog) if on the same chromosome (left), in the dog genome. In addition, genes in each example are all amplified in the human tumors; however, in the dog tumors, only DCGs [GADD45A and SLC2A1, both of which are already known cancer driver genes (22, 23)] are amplified, whereas PCGs (genes shown in red, none of which has been reported in the literature to be cancer driver genes) remain neither amplified nor deleted.