Figure 1. GPS profiling identifies host proteins altered during C. trachomatis infection.
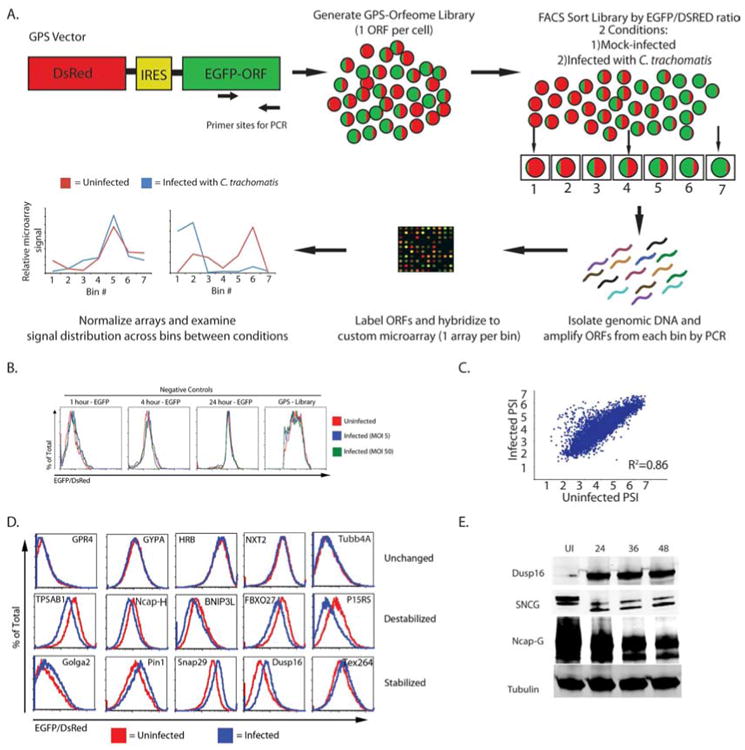
A) Schematic representation of the GPS screen.
B) Histograms show the EGFP/DsRed ratio for cells expressing GFP for varying half-life and the entire GPS library. The cell lines were mock-infected (red line) or infected with C. trachomatis at an MOI of 5 (blue line) or 50 (green line) for 24 hours and analyzed by flow cytometry.
C) Scatter plot for the PSI for each probe in the screen under the two conditions (mock-infected vs. infected).
D) A subset of candidates that were tested by expressing the host gene in HEK293T cells. Cells were mock-infected (red line) or infected with C. trachomatis (blue line) for 24 hours and evaluated using flow cytometry. Shown is a representative plot from three independent experiments.
E) HEK293T cells were mock-infected or infected with C. trachomatis for the indicated amount of time. Cells were analyzed by immunoblot for the expression of the experimental proteins (Dusp16, SNCG, and NCAP-G) as well as beta-tubulin (loading control). Blots are representative of at least 3 independent experiments. See also Figure S1, Table S1, S2, and S4.
