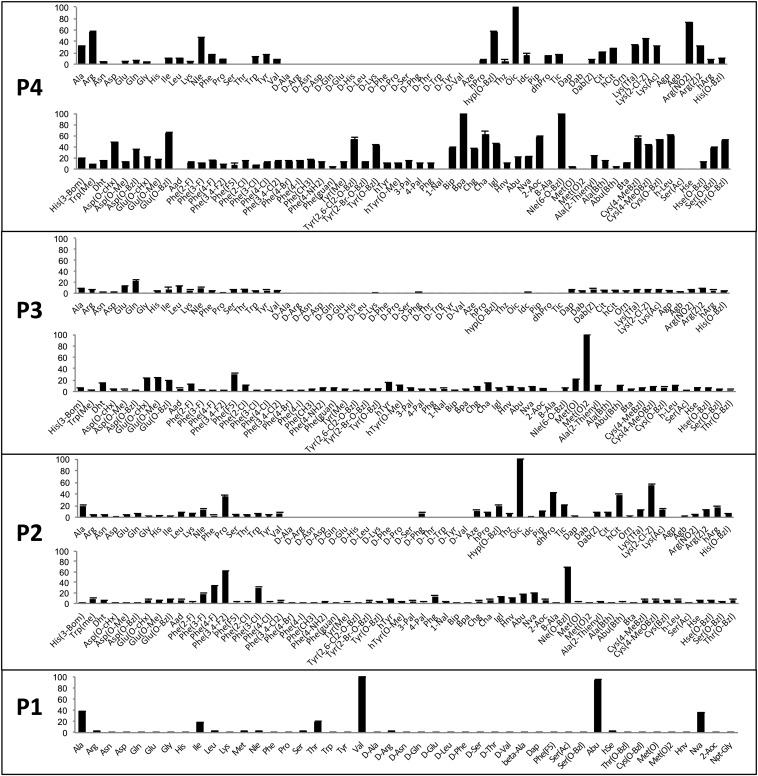
Fig. 2.
Preferences at the P4–P1 subsites of NE. The P4, subsite preference of NE was determined using combinatorial substrate libraries of the general structure Ac-P4-X-X-Ala-ACC, where P4 represents a natural or unnatural amino acid and X represents an isokinetic mixture of natural amino acids (Cys and Met were omitted because of a problem with oxidation; Nle was used instead of Met). The P3 and P2 preferences were determined in a similar way. The P1 subsite preference was determined using individual substrate libraries of the general structure Ac-Ala-Ala-Pro-P1-ACC, where P1 is a natural or unnatural amino acid. Abbreviated amino acid names (SI Appendix, Fig. S2) are shown on the x axis. The y axis displays the average relative activity expressed as a percentage of the best amino acid. Error bars represent the SD (n = 3).