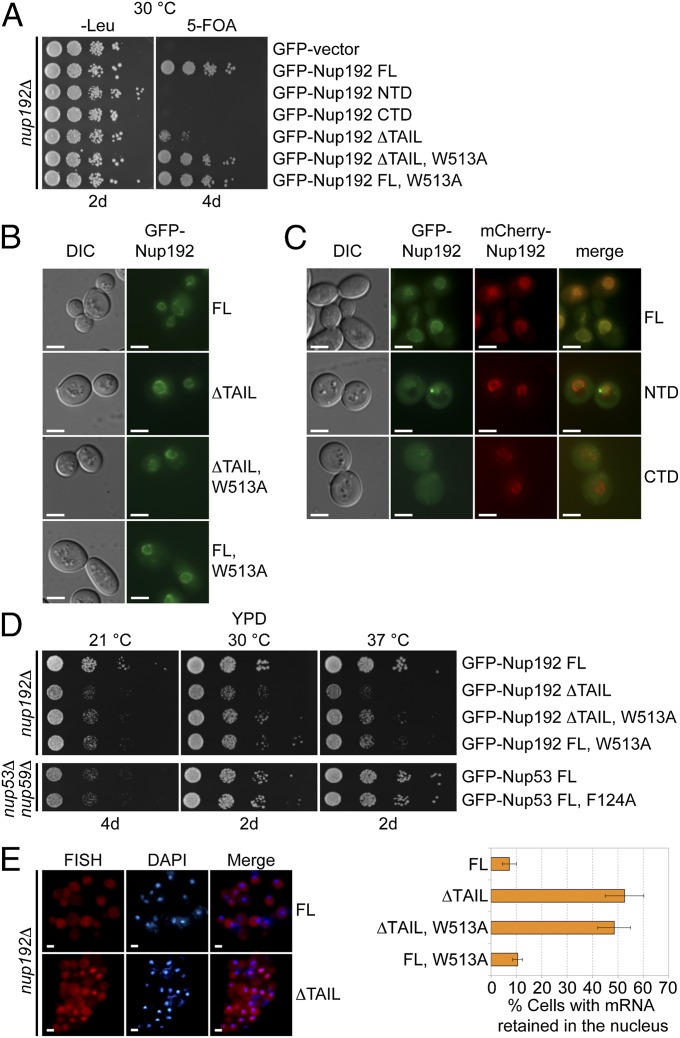
Fig. 6.
In vivo analyses of Nup192 and Nup53 mutants in S. cerevisiae. (A) The nup192Δ shuffle strain containing an mCherry-Nup192 cover plasmid (URA3) was transformed with control or GFP-Nup192 variants [leucine (Leu)]. Growth was followed on synthetic dextrose complete (SDC)-Leu and 5-fluoroorotic acid (5-FOA)/SDC plates for the indicated times and temperatures. FL, full-length. (B and C) In vivo localization of GFP-Nup192 variants in the nup192Δ strain visualized by fluorescence and differential interference contrast (DIC) microscopy. (C) Cells still carry the mCherry-Nup192 full-length plasmid. (D) Growth analysis of nup192Δ and nup53Δnup59Δ strains. Cells were spotted on yeast extract peptone dextrose (YPD) plates and grown for 2–4 d at the indicated temperatures. (E) mRNA export assay of GFP-Nup192 variants. (Left) Representative images of WT GFP-Nup192– (Upper) and GFP-Nup192∆TAIL– (Lower) complemented nup192Δ cells are shown. (Right) Quantification of nuclear poly(A) mRNA retention is shown. Error bars indicate SDs derived from six independent images, each containing ∼100 cells. (Scale bars: 5 μm.)