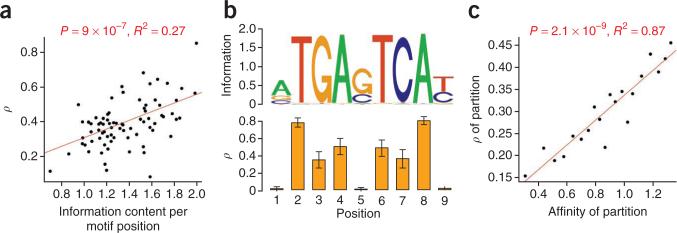
Figure 3.
Information content, binding affinity and selection. (a) Information content per motif position versus estimates of ρ (the fraction of sites under selection) for the 78 transcription factors analyzed in our study. (b) Motif logo for JUND (top) and position-specific estimates of ρ (bottom). Error bars, 1 s.e.m. Notice that positions with high information content tend to be under selection, and positions with low information content tend not to be under selection. This relationship holds for some but not all transcription factors. (c) Predicted binding affinity versus ρ. All binding sites were partitioned into 20 equally sized bins by predicted binding affinity, and ρ was estimated separately for each partition using INSIGHT. Additional details are given in the supplementary Note.