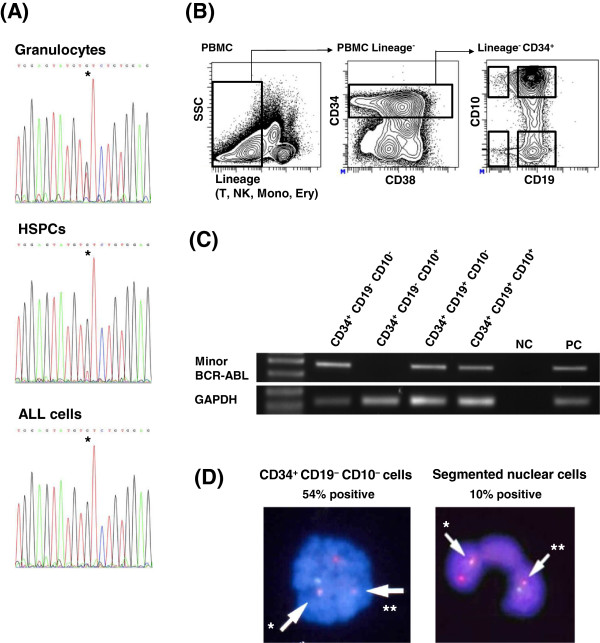
Figure 1.
Analysis of the molecular based clonal architecture. (A) Sequencing of JAK2. Granulocytes and FACS-sorted lineage-CD34+ cells (HSPCs) and CD34+CD19+ B-ALL cells were analyzed. The JAK2-V617F mutation was not detected in B-ALL cells. Asterisk indicates nucleotide 1849 of JAK2. Lineage markers included CD2, CD3, CD4, CD7, CD8, CD10, CD11b, CD14, CD19, CD20, CD56 and CD235. (B) FACS analysis and sorting of PBMCs at diagnosis. The gating strategy to isolate four populations is shown. Lineage markers included CD2, CD3, CD4, CD7, CD8, CD11b, CD14, CD56 and CD235. (C) RT-PCR analysis for each population (gated in (B)). Plasmids containing the amplified region of minor BCR-ABL or GAPDH were used as positive controls (PC). Distilled water was used as the negative control (NC). The left lane shows the size marker. The Minor BCR-ABL transcript was also detected in CD34+CD19-CD10- cells. (D) FISH analysis of BCR-ABL utilizing probes of Vysis LSI ASS-ABL for 9q34 (red) and Vysis LSI BCR for 22q11.2 (green). One red-green fusion signal specified by the arrow* indicates the presence of BCR-ABL. One smaller red signal specified by the arrow** indicates the remaining part of 9q34. Translocation t(9;22) was detected in both CD34+CD19-CD10- cells at diagnosis and in segmented nuclear cells four days after the initiation of dasatinib treatment.