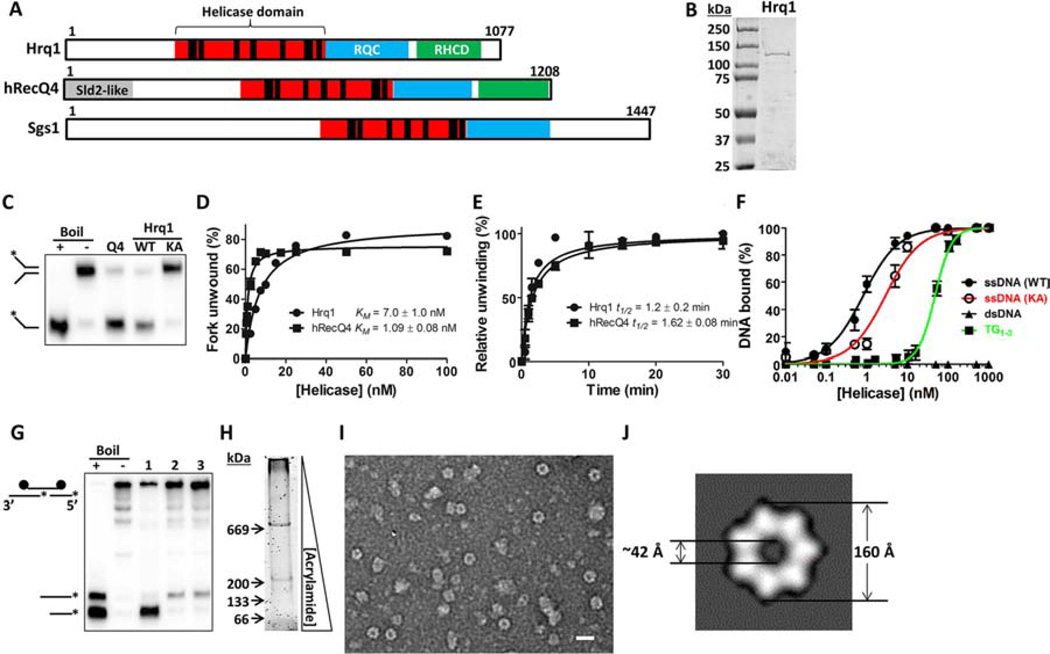
Figure 1. Purified Hrq1 is an active 3’–5’ helicase.
(A) Domain schematics of Hrq1, hRecQ4, and Sgs1. The amino acid length of each is given on the right. The black bars in the helicase domain correspond to conserved ATPase/helicase motifs. RQC: RecQ C-terminal domain. RHCD: RecQ4/Hrq1-conserved domain. Sld2-like: portion of hRecQ4 homologous to S. cerevisiae Sld2. (B) Coomassie-stained gel of purified S. cerevisiae Hrq1. The expected molecular weight is ~130 kDa. (C) hRecQ4 (Q4) and Hrq1 (WT) (both 50 nM) unwind a fork substrate; 100 nM Hrq1-KA (KA) does not. (D) Hrq1 and hRecQ4 unwind the fork with similar apparent KMs ([protein] necessary to unwind 50% of the DNA). (E) The rates of fork unwinding by 50 nM Hrq1 and hRecQ4 were similar (t1/2 = time necessary to unwind 50% of the DNA). (F) Hrq1 and Hrq1-KA bind ssDNA by gel shifts. Hrq1 also preferentially bound ss- vs. dsDNA, as well as telomeric repeat ssDNA (TG1–3). (G) Directionality of Pif1 (lane 1), hRecQ4 (lane 2), and Hrq1 (lane 3) unwinding. The fastest migrating band corresponds to 5’-3’ unwinding; the slower migrating band indicates 3’–5’ activity. (H) Sypro Orange-stained native gradient PAGE gel of Hrq1. (I) TEM image of negative-stained Hrq1; white bar = 200 Å. (J) Two-dimensional reconstruction of the Hrq1 heptamer. The inner and outer diameters of the ring are shown. All gel images are representative of ≥3 independent experiments, plotted data represents the average of ≥3 independent experiments, and error bars correspond to the standard deviation. See also Figures S1 and S5.