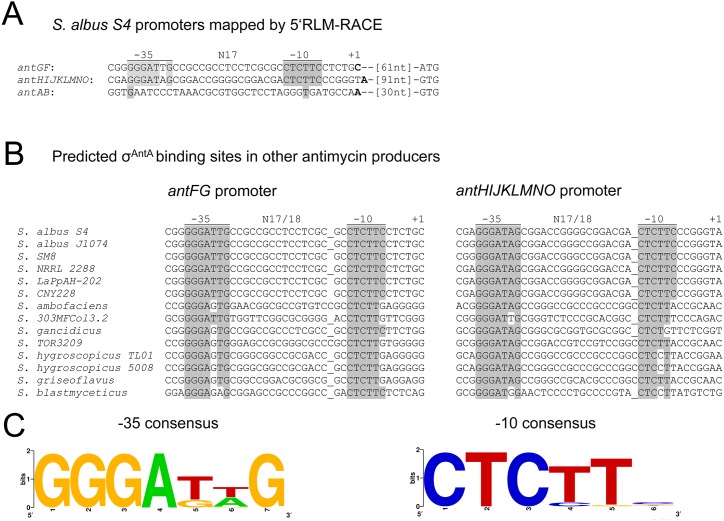
Figure 7. Identification of σAntA promoter motifs.
(A) The −10 and −35 motifs at the σAntA-target promoters of antFG and antHIJKLMNO are nearly 100% identical and display zero nucleotide identity with the promoter region of antAB. Shared identity is indicated by grey shading. The nucleotides mapped by 5′RLM-RACE is denoted by +1 and are shown in bold face (B) Conservation of the S. albus S4 antFG and antHIJKLMNO promoter elements in other antimycin-producing Streptomyces species. Conservation between the experimentally determined promoter region of S. albus S4 and the putative promoter regions of other antimycin producers is indicated by grey shading. (C) Consensus sequence for the −35 and −10 promoter elements recognised by σAntA displayed as a WebLogo (Crooks et al., 2004). Below are the full strain names and accession numbers for antimycin-producing strains: S. ambofaciens ATCC 23877 (AM238663), S. blastmyceticus NBRC 12747 (AB727666), S. gancidicus BKS 13-15 (AOHP00000000), S. griseoflavus T4000 (ACFA00000000), S. hygroscopicus subsp. jinggangensis 5008 (NC_017765), S. hygroscopicus subsp. jinggangensis TL01 (NC_020895), Streptomyces sp. 303MFCol5.2 (ARTR00000000), Streptomyces sp. TOR3209 (AGNH00000000), S. albus S4 (CADY00000000), S. albus J1074 (NC_020990), Streptomyces sp. SM8 (AMPN00000000), Streptomyces sp. NRRL2288 (JX131329), Streptomyces sp. LaPpAH-202 (ARDM00000000), Streptomyces sp. CNY228 (ARIN00000000).