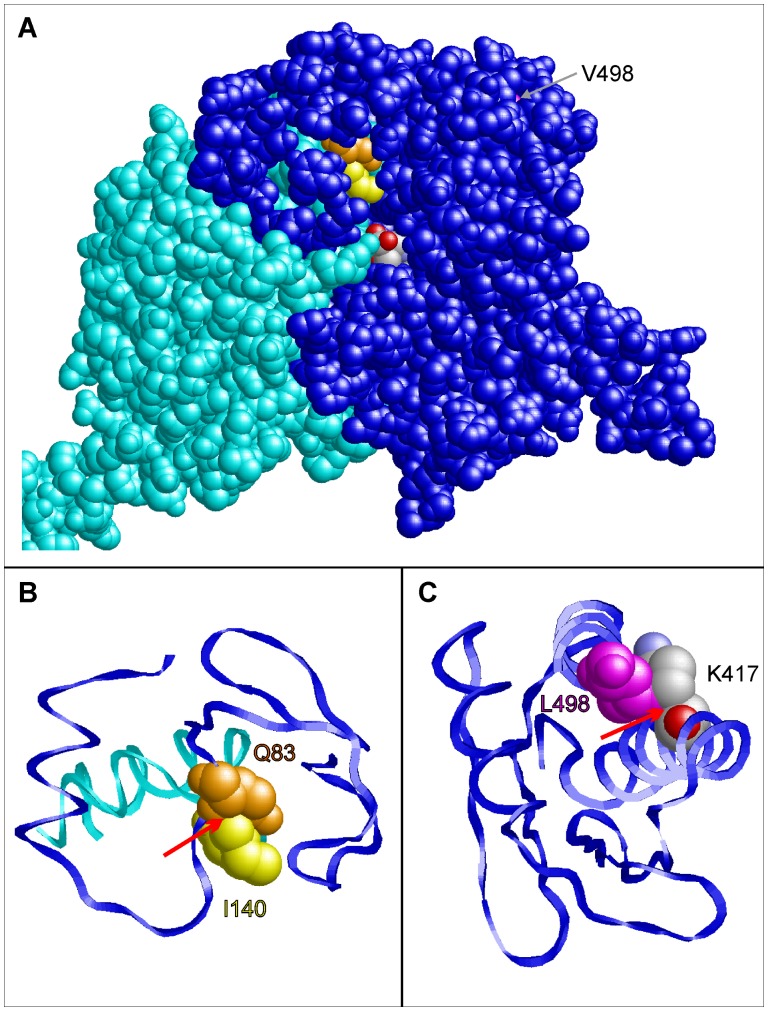
Figure 3. Prediction of the phenotypic effects of the coding SNPs rs37369 (p.Val140Ile) and rs16899974 (p.Val498Leu) in AGXT2 using structural information from in silico homology modeling.
(A) Three-dimensional model of the AGXT2 (V140I) dimer showing the subunits A (blue) and B (cyan) in space-filled presentation. Residues Q83 (chain A), I140 (chain B), and V498 (chain A) are colored in orange, yellow and magenta, respectively. The β-aminoisobutyrate substrate of subunit A is colored according to the atom types. This view shows that V498 is buried in the interior of the molecule, whereas I140 of chain B is located close to the substrate binding site of subunit A. (B) Enlargement of the site of the V140I mutation. Residues 60–120 of chain A (blue) and residues 120–150 of chain B (cyan) are shown in ribbon presentation. A red arrow denotes a clash between the side chains of I140 (chain B; yellow) and Q83 (chain A; orange) that is not observed for V140 in the wild-type. (C) Enlargement of the C-terminal residues 400–500 showing the effect of a V498L mutation. The larger side chain of L498 forms steric clashes with the methylene groups of the K417 side chain (denoted by a red arrow).