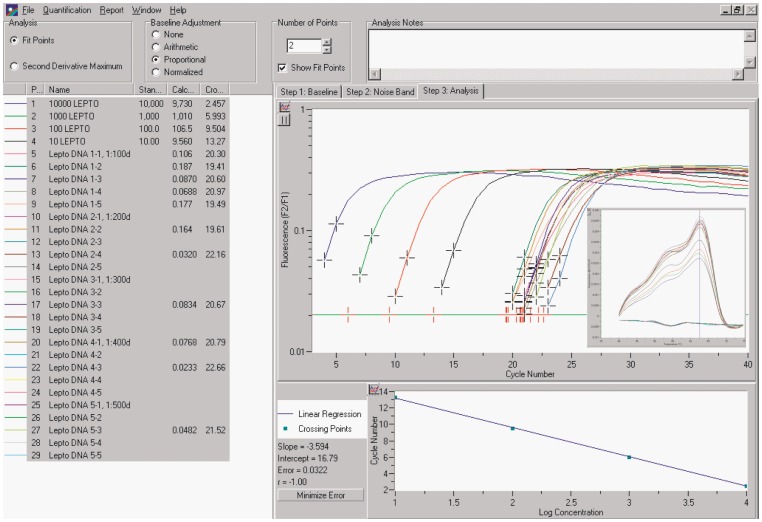
Figure 2. Detection limit of the Leptospira spp. lig FRET-PCR.
For use as quantitative standard in the lig PCR, DNA from a pure culture of a wild-type isolate of Leptospira interrogans serovar Pomona was extracted and quantified by PicoGreen DNA fluorescence assay (Invitrogen, Karlsbad, CA, USA). The number of genomes was calculated assuming a Leptospira spp. genome size of 4.63 Mb (MW = 3.06×109 Dalton), which was further used to determine the sensitivity of the lig FRET-PCR by amplification of logarithmic dilutions of this standard. To calculate the actual number of lig gene targets per leptospiral genome, the 10-copy standard DNA was further serially diluted from 1∶100-fold (0.1 genomes/PCR) to 1∶500-fold (0.02 genomes/PCR), and 5 aliquots of each dilution step were subjected to PCR detection. This approach yielded a Poisson distribution of positive and negative amplification reactions, with fewer of the 5 aliquots positive at higher dilutions. Logistic regression analysis indicated a 50% probability of a positive PCR at a PCR input of 0.0476 genomes. The complete separation between positive and negative reactions indicates a robust PCR methodology that reliably amplifies single target copies. Thus, we calculated from the mean of 0.5 targets per 0.0476 genomes an approximate number of 10.5 targets per leptospiral genome. The insert of the melting curves shows that positive amplification of leptospiral DNA is associated with the presence of a specific melting point (T m = 62.5°C) which does not exist in the samples free of leptospiral DNA.