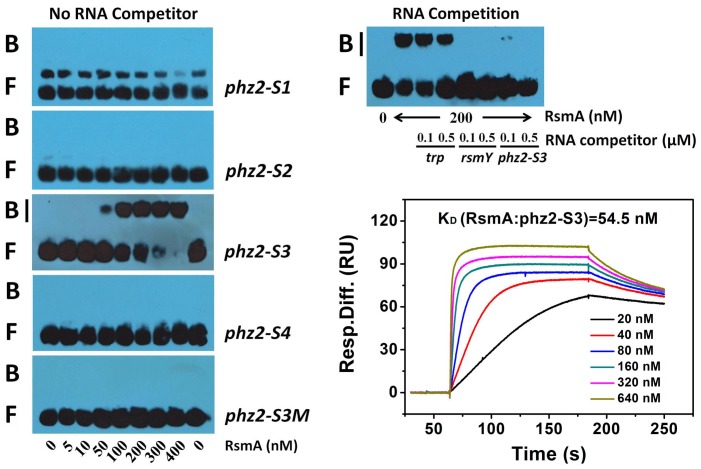
Figure 5. Gel mobility shift assays for RsmA protein direct binding to the 5′-UTR of the phz2 transcript.
3′ end-labeled WT phz2-S1, phz2-S2, phz2-S3, phz2-S4 and mutant phz2-S3M transcripts (1 nM) were incubated with RsmA (concentration indicated at the bottom of each lane). Positions of bound (B) and free (F) phz2 leader RNA are marked. For an RNA competition assay, labeled phz2-S3 RNA was incubated with RsmA ±100- or 500-fold excess of non-specific (trp) and specific (rsmY and phz2-S3) competitor RNA. The concentrations of RsmA and competitor RNA were shown at the bottom of the corresponding lanes. The equilibrium binding constant (KD) of RsmA binding to WT phz2-S3 was calculated to be 54.5 nM using surface plasmon resonance assays.