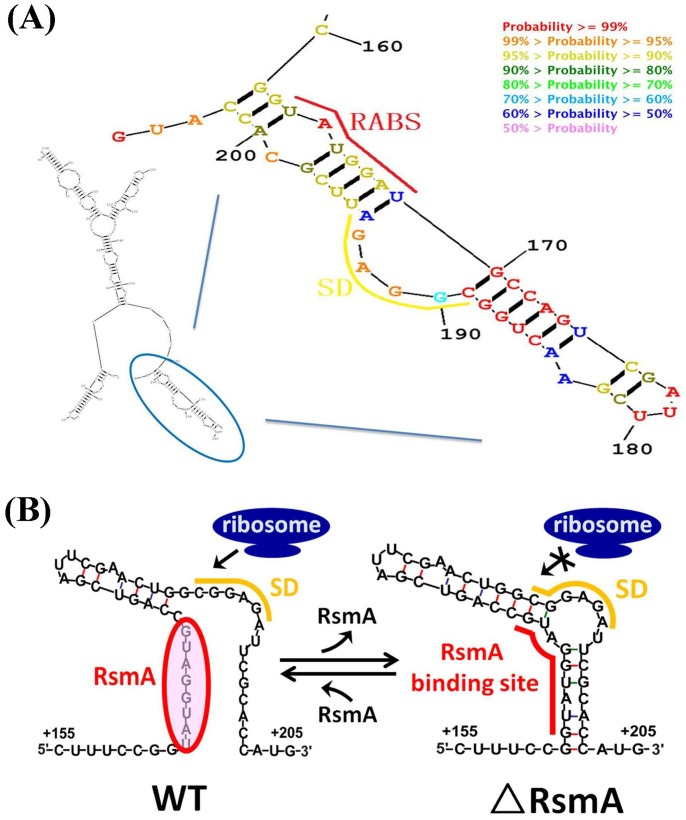
Figure 7. Structure prediction of the phz2 transcript and putative model of RsmA mediated activation of phz2 translation.
(A) A predicted 5′-UTR secondary structure from +160 to +205 nucleotides of the phz2 transcript generated by RNA structure. (B) A putative direct activation model of the phz2 transcript mediated by RsmA in P. aeruginosa M18. In the rsmA-deleted mutant (right), base-paired nucleotides between the SD with its flanking sequence and the RABS resulted in a relatively stable stem-loop structure, which prevented ribosome access and translation initiation. However, in the WT strain (left), the loose stem-loop structure caused by RsmA binding resulted in easy access to the ribosome and translation activation of the phz2 transcript. RABS: RsmA binding site.