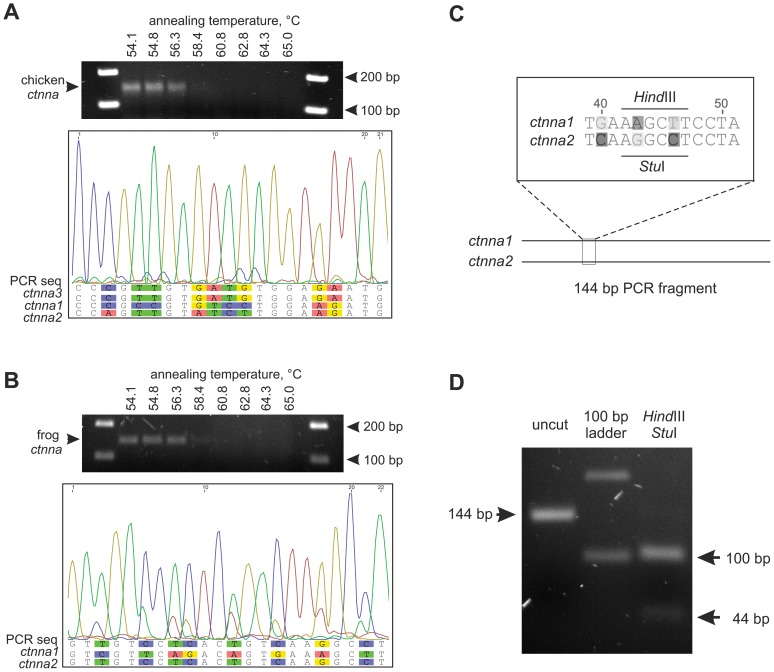
Figure 4. Experimental evidence that the clawed frog genome does not contain a ctnna3 ortholog.
(A) The degenerate primers a3-F and a3-R (corresponding to the conserved last coding exon of ctnna3 in vertebrates, see Fig. S3) were used to amplify corresponding fragments of the ctnna genes from the chicken genomic DNA. PCR product of the predicted size (about 150 bp) was observed using annealing temperatures from 54.1°C to 56.3°C. Sequencing of the PCR fragment (with the same primers) revealed spectra corresponding mainly to chicken ctnna3. Minor peaks corresponding to chicken ctnna1 PCR product are slightly shifted to the right. (B) The same primers were used to amplify corresponding fragments of the ctnna genes from the clawed frog genomic DNA. See also Fig. S4. Sequencing of the PCR fragment revealed spectra corresponding to the frog ctnna1 and ctnna2 genes only. Shown is a part of the sequence spectrum obtained with a3-F primer. (C) Schematic drawing of the experimental strategy. A PCR of X. tropicalis genomic DNA using degenerate ctnna3 primers is expected to amplify 144 bp fragments of frog ctnna1 and ctnna2 that contain HindIII and StuI restriction enzyme sites, respectively. (D) Arrow on the left points at the 144 bp PCR product obtained from the frog genomic DNA. Sequencing of this band is shown in B. Arrows on the right indicate the diagnostic HindIII/StuI fragments of the PCR product verifying that the product is solely composed of the predicted ctnna1 and ctnna2 fragments.