Figure 6.
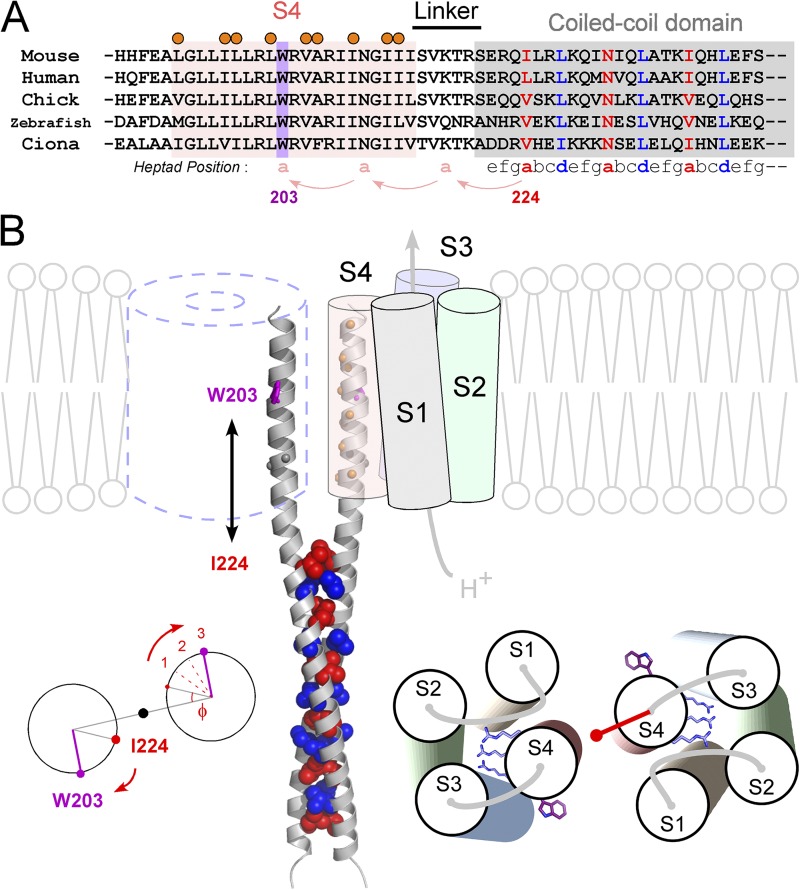
Molecular architecture of the dimeric Hv. (A) Sequence alignment of S4 and the C-terminal cytoplasmic coiled-coil domain of Hv from various species. Positions of the coiled-coil heptad repeat (abcdefg) are indicated below the alignment (Lupas, 1996; Burkhard et al., 2001). Coiled-coil residues occupying hydrophobic “a” and “d” positions are denoted by blue and red, respectively. The conserved W203 in S4 is highlighted in violet. Positions showing high cross-linking ratios (Fig. 3) are marked by orange circles. (B) A model of the Hv dimer based on the experimental data and the crystal structures of the coiled-coil domain of Hv (Fujiwara et al., 2012) and VSD of Kv (Long et al., 2005). Spheres depicting the core residues of “a” (red) and “d” (blue) positions of the coiled-coil on ribbon backbones (gray) are shown. W203 and orange balls depicting the residues showing the high cross-linking ratio are shown on the continuous helix ribbon backbones from the coiled-coil. N210 and Gly211 (gray balls) are also highlighted because of an alternative model in which a continuous helix might be broken here. Calculation of the phase angle at W203, which is 21 residues, 3-heptad, upstream of I224 (left). The model for the helix orientation of the VSD based on the cross-linking (right). Three conserved Arg’s and W203s are shown as sticks. The red vector depicts the sum vector of Fig. 3 D. In this study, the transmembrane region, i.e., the S4 segment, was defined based on the sequence alignment with the Kv family and the hydrophobicity profile of the S4 segment. The observation that the gradual shift of the threshold by decrease of the linker length (greater than or equal to −5 A.A.) (Fig. 1 B) might be caused by the hydrophobic mismatch between the lipid membrane and the coiled-coil domain, of which surface residues are hydolophilic, in addition to the interpretation of the restriction of the S4 movement by the coiled-coil.