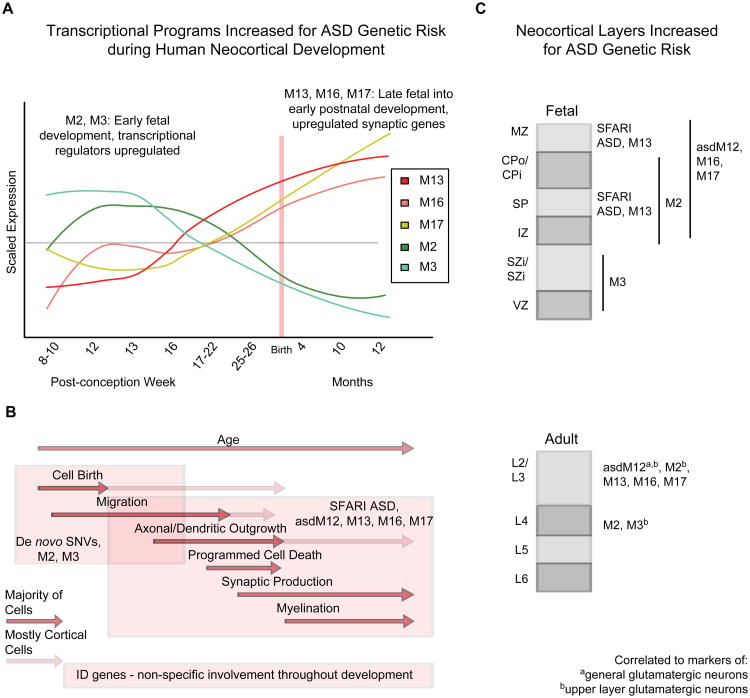
Figure 7. Summary of findings and model for effects of ASD implicated gene sets.
(A) ASD risk genes from multiple sources were enriched in five co-expression modules throughout development, M2, M3, M13, M16, and M17.
(B) Early transcriptional regulators in M2/M3 are enriched for RDNVs, while the later expressed synaptic genes are associated with previously studied ASD genes (Biological processes time periods adopted from Andersen, 2003).
(C) ASD genes are most consistently associated with laminae containing post-mitotic neurons during early fetal development (broadly in IZ, SP, CPo/CPi, and MZ) and superficial layers in adult (L2-4). Multiple modules are also strongly associated with markers of upper layer glutamatergic neurons in adult cortex, suggesting many ASD genes preferentially affect these cell types.
B) and C) also summarize that ID genes are largely distinct from ASD genes in both developmental trajectory and neocortical layer enrichment.
Both figures A and B correspond to the same time scale as marked by the axis on the plot in (A). We summarize the strongly enriched findings, but note that weaker enrichment for other patterns exists that may be important for subsets of ASD. Individual genes can be prioritized for biological validation using a combination of network position, bioinformatic scores, and the biological context highlighted here, as discussed in the text and shown in Table S4.