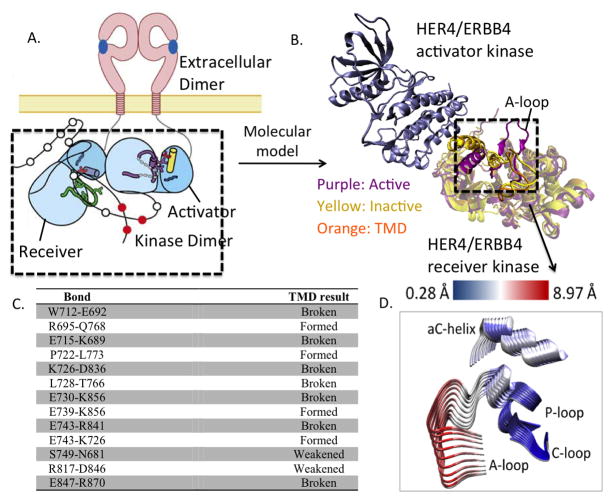
Figure 1.
(A) Schematic of ligand-mediated dimerization of extracellular and intracellular domains in ErbB kinases. (B) Molecular model of the asymmetric dimer of the HER4 kinase domain. (C) List of bonds formed or broken during the course of the TMD simulation of HER4/ErbB4 activation. (D) Principal Component Analysis (PCA) of the HER4 dimer. Superimposing several frames from the each PCA trajectory, we depict conformational fluctuations along the first principal component. For clarity, only the key regions, namely, A-loop, P-loop, C-loop, and αC helix of the receiver kinase are depicted. That is, we take the principal component (which is a 3N dimensional vector) and only show its projection relevant a sub-dimensional vector involving the alpha-C-helix, A-loop, P-loop, N-loop atoms of the kinase undergoing activation. The structures are colored according to the RMSD, where blue regions indicate small fluctuations and red regions indicate larger fluctuations. The fluctuations are quantified according to the scale bar at the top.