Abstract
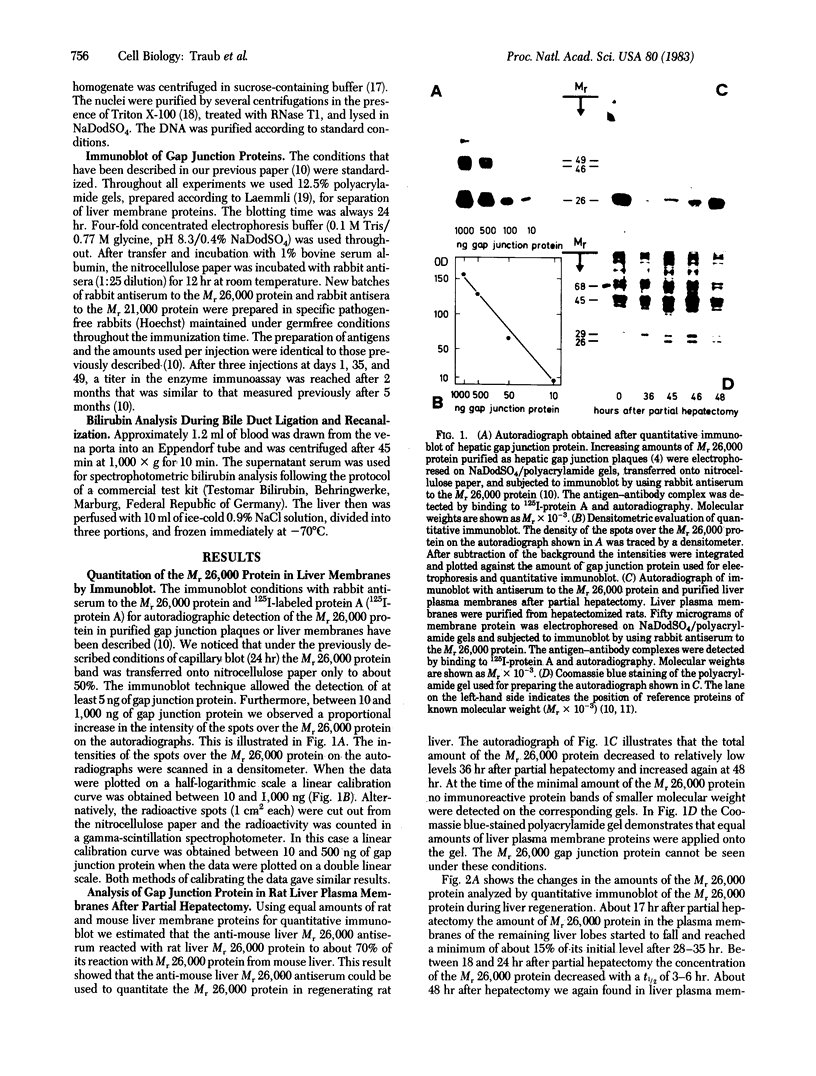
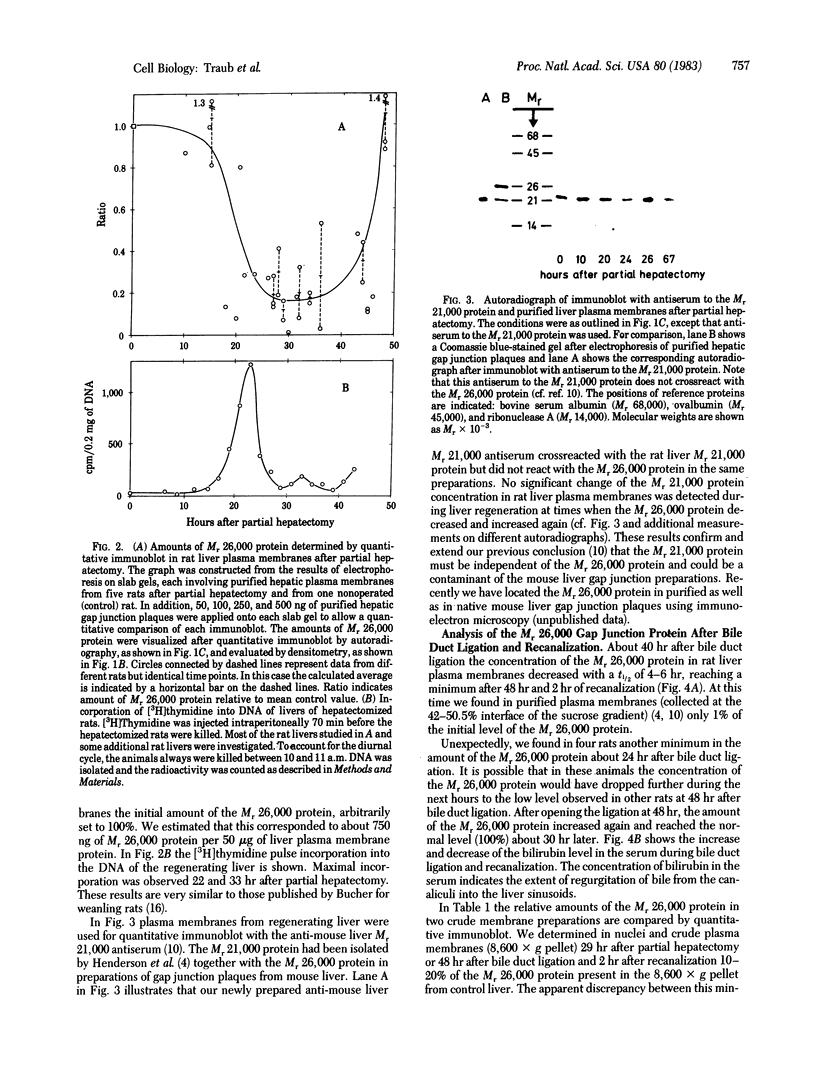
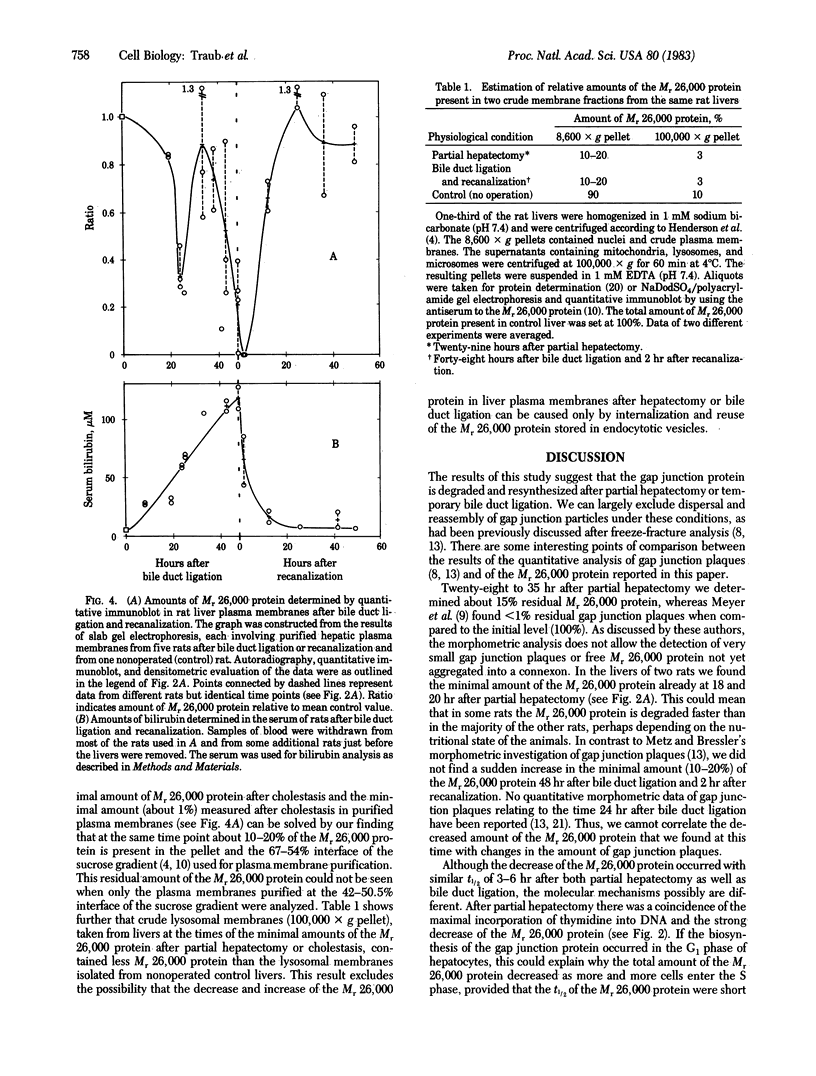
Changes in the total amount of the gap junction protein (Mr 26,000) after partial hepatectomy or bile duct ligation and recanalization were investigated in rat liver membranes by quantitative immunoblot with rabbit antiserum to the Mr 26,000 protein. The loss and reappearance of the Mr 26,000 protein roughly paralleled loss and reappearance of gap junction plaques analyzed previously under similar physiological conditions by freeze-fracture of hepatocyte surfaces. The total amount of the hepatic Mr 26,000 protein in liver plasma membranes and the total area of the hepatocyte surface occupied by gap junction plaques appeared to be proportional under these conditions. However, at the minimum, 28-35 hr after partial hepatectomy we still find about 15% of the Mr 26,000 protein, in contrast to <1% of gap junction plaques, determined by morphometric analysis. This discrepancy is probably due to the fact that very small gap junction plaques, single connexons, and free Mr 26,000 gap junction subunits are missed by the morphometric analysis. At the times of the minimal amount of the Mr 26,000 protein in hepatic plasma membranes after partial hepatectomy or bile duct ligation we found that crude hepatic lysosomal membranes of these rats contained less Mr 26,000 protein than lysosomal membranes of nonoperated control animals. Thus, we conclude that the decrease and increase of the total amount of the Mr 26,000 protein cannot be explained only by dispersal and reuse of gap junction subunits but are largely due to degradation and resynthesis of the Mr 26,000 protein. No significant change in the amount of the Mr 21,000 protein that had been isolated with gap junction plaques was observed in liver plasma membranes after partial hepatectomy. This confirms our previous conclusion that the Mr 26,000 and Mr 21,000 proteins are independent of each other.
Keywords: quantitative immunoblot, bile duct ligation, recanalization, cell-cell communication, metabolic cooperation
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BUCHER N. L. REGENERATION OF MAMMALIAN LIVER. Int Rev Cytol. 1963;15:245–300. doi: 10.1016/s0074-7696(08)61119-5. [DOI] [PubMed] [Google Scholar]
- Blobel G., Potter V. R. Nuclei from rat liver: isolation method that combines purity with high yield. Science. 1966 Dec 30;154(3757):1662–1665. doi: 10.1126/science.154.3757.1662. [DOI] [PubMed] [Google Scholar]
- Fallon R. F., Goodenough D. A. Five-hour half-life of mouse liver gap-junction protein. J Cell Biol. 1981 Aug;90(2):521–526. doi: 10.1083/jcb.90.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finbow M., Yancey S. B., Johnson R., Revel J. P. Independent lines of evidence suggesting a major gap junctional protein with a molecular weight of 26,000. Proc Natl Acad Sci U S A. 1980 Feb;77(2):970–974. doi: 10.1073/pnas.77.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greim H., Trülzsch D., Roboz J., Dressler K., Czygan P., Hutterer F., Schaffner F., Popper H. Mechanism of cholestasis. 5. Bile acids in normal rat livers and in those after bile duct ligation. Gastroenterology. 1972 Nov;63(5):837–845. [PubMed] [Google Scholar]
- Henderson D., Eibl H., Weber K. Structure and biochemistry of mouse hepatic gap junctions. J Mol Biol. 1979 Aug 5;132(2):193–218. doi: 10.1016/0022-2836(79)90391-7. [DOI] [PubMed] [Google Scholar]
- Hertzberg E. L., Anderson D. J., Friedlander M., Gilula N. B. Comparative analysis of the major polypeptides from liver gap junctions and lens fiber junctions. J Cell Biol. 1982 Jan;92(1):53–59. doi: 10.1083/jcb.92.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper M. L., Subak-Sharpe J. H. Metabolic cooperation between cells. Int Rev Cytol. 1981;69:45–104. doi: 10.1016/s0074-7696(08)62320-7. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Loewenstein W. R. Junctional intercellular communication: the cell-to-cell membrane channel. Physiol Rev. 1981 Oct;61(4):829–913. doi: 10.1152/physrev.1981.61.4.829. [DOI] [PubMed] [Google Scholar]
- Metz J., Aoki A., Merlo M., Forssmann W. G. Morphological alterations and functional changes of interhepatocellular junctions induced by bile duct ligation. Cell Tissue Res. 1977 Aug 26;182(3):299–310. doi: 10.1007/BF00219766. [DOI] [PubMed] [Google Scholar]
- Metz J., Bressler D. Reformation of gap and tight junctions in regenerating liver after cholestasis. Cell Tissue Res. 1979 Jun 27;199(2):257–270. doi: 10.1007/BF00236137. [DOI] [PubMed] [Google Scholar]
- Meyer D. J., Yancey S. B., Revel J. P. Intercellular communication in normal and regenerating rat liver: a quantitative analysis. J Cell Biol. 1981 Nov;91(2 Pt 1):505–523. doi: 10.1083/jcb.91.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh H. Freeze replica observations on the junction structures in the guinea-pig liver after bile duct ligation and recanalization. Arch Histol Jpn. 1981 Sep;44(4):345–367. doi: 10.1679/aohc1950.44.345. [DOI] [PubMed] [Google Scholar]
- Nicholson B. J., Hunkapiller M. W., Grim L. B., Hood L. E., Revel J. P. Rat liver gap junction protein: properties and partial sequence. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7594–7598. doi: 10.1073/pnas.78.12.7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng C. S., Teng C. T., Allfrey V. G. Studies of nuclear acidic proteins. Evidence for their phosphorylation, tissue specificity, selective binding to deoxyribonucleic acid, and stimulation effects on transcription. J Biol Chem. 1971 Jun 10;246(11):3597–3609. [PubMed] [Google Scholar]
- Traub O., Janssen-Timmen U., Drüge P. M., Dermietzel R., Willecke K. Immunological properties of gap junction protein from mouse liver. J Cell Biochem. 1982;19(1):27–44. doi: 10.1002/jcb.240190104. [DOI] [PubMed] [Google Scholar]
- Yancey S. B., Easter D., Revel J. P. Cytological changes in gap junctions during liver regeneration. J Ultrastruct Res. 1979 Jun;67(3):229–242. doi: 10.1016/s0022-5320(79)80024-6. [DOI] [PubMed] [Google Scholar]
- Yancey S. B., Nicholson B. J., Revel J. P. The dynamic state of liver gap junctions. J Supramol Struct Cell Biochem. 1981;16(3):221–232. doi: 10.1002/jsscb.1981.380160303. [DOI] [PubMed] [Google Scholar]
- Yee A. G., Revel J. P. Loss and reappearance of gap junctions in regenerating liver. J Cell Biol. 1978 Aug;78(2):554–564. doi: 10.1083/jcb.78.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]