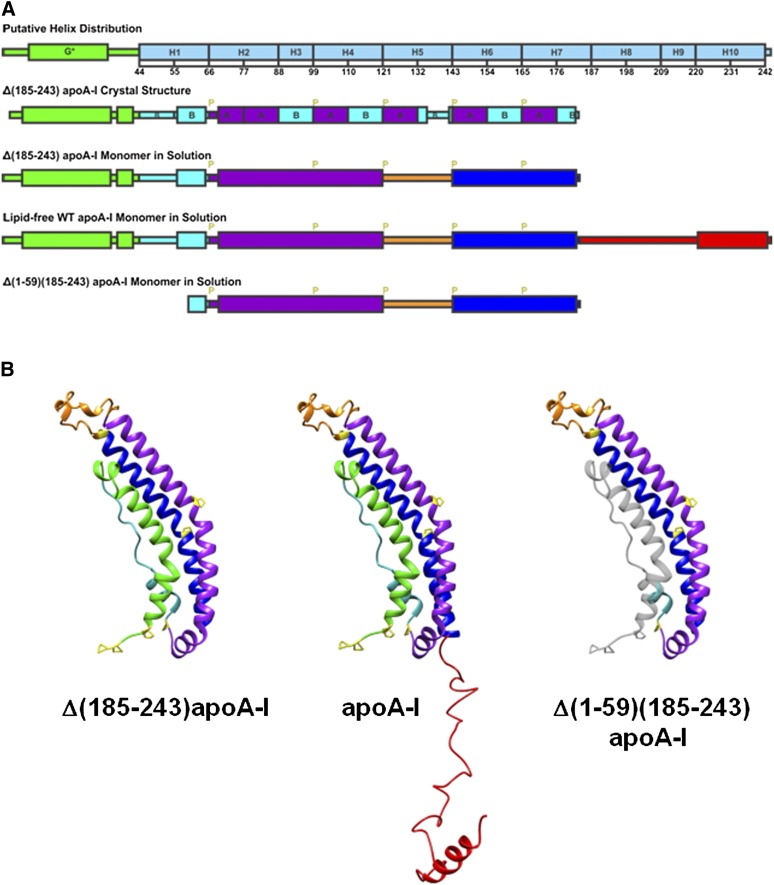
Fig. 6.
A: Illustrations of the helix distribution from sequence analysis (10), Δ(185-243)apoA-I crystal structure (19), and the predicted monomer secondary structures. G*stands for the type G* AαH, H stands for the predicted Helix 1 to 10 (10), B and A stand for the B and A repeats of AαHs of apoA-I (19), and P stands for proline. B: Predicted monomer structures of Δ(185-243)apoA-I, full-length apoA-I, and Δ(1-59)(185-243)apoA-I in solution based on the crystal structure of the Δ(185-243)apoA-I dimer (19). The full-length apoA-I consists of an N-terminal α-helix bundle domain (residues 1-184), and a less organized C-terminal domain (residues 185-243). The four parts of the N-terminal α-helix bundle domain are the exon 3 encoded N-terminal region (residues 1-43, in green), helix 1 (residues 44-65, in cyan), helix 2-4 (residues 66-120, in purple), and helix 6-7 (residues 143-184, in blue), which folds back on the bundle by a hinge at helix 5 (residues 121-142, in orange). The C-terminal domain (residues 185-243, in red) extends away from the bundle and has an AαH at the end (residues 220-241). Proline residues are labeled in yellow in the monomer structures. The domain distributions are colored accordingly. The missing residues 1-59 from the Δ(1-59)(185-243)apoA-I structure are shown in gray for comparison purposes.