Table 1.
Comparison of a GATK and SAMtools’s Multisample Calling Pipeline.
| Step | GATK | SAMtools |
|---|---|---|
| [Calculating Genotype Likelihoods] For each individual, at each site, the likelihoods for 10 possible genotypes (AA,GG,CC,TT,AC,AG,AT,CG,CT,GT) are computed based on aligned reads. | Independent errors assumed. | Dependent errors assumed. |
[SNP calling] At each site, determine whether a site is polymorphic based on posterior probabilities of nonreference allele counts P(Xa|D,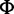 ) where ) where 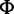 is an expected SFS under the standard model and D is aligned reads. is an expected SFS under the standard model and D is aligned reads. |
A site is polymorphic if a 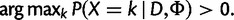
|
A site is polymorphic if 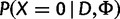 < cutoff (default = 0.5). < cutoff (default = 0.5). |
| [Genotype Calling] If a site is considered polymorphic, the maximum a posteriori genotype is assigned to each individual. | At each site, the same genotype prior probabilities are used: P(AA) = 1 − 3θ/2 P(Aa) = θ P(aa) = θ/2, where θ is an expected heterozygosity (default = 0.001) | At each site, genotype prior probabilities are computed based on the estimated nonreference allele frequency q and assuming Hardy–Weinberg equilibrium: P(AA) = p2 P(Aa) = 2pq P(aa) = q2 |
aX denotes nonreference allele counts in a sample of n individuals.
